Abstract
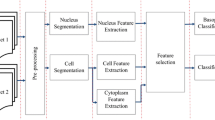
Blood is composed of white blood cells, red blood cells, and platelets. Segmentation of the blood smear cells and extraction of features of the cells is essential in the field of medicine. Acute lymphoblastic leukemia is a form of blood cancer caused due to the abnormal increase in the production of immature white blood cells in the bone marrow. It mostly affects the children below 5 years and adults above 50 years of age. Due to the late diagnosis and cost of the devices used for the determination, the mortality rate has increased drastically. Flow cytometry technique that performs automated counting fails to identify the abnormal cells. Manual recount performed using hemocytometer are prone to errors and are imprecise. The proposed work aims to survey different computer-aided system techniques used to segment the blood smear image. The primary objective here is to derive knowledge from the different methodologies used for extracting features from white blood cells and develop a system that would accurately segment the blood smear image by overcoming the drawbacks of the previous works. The objective mentioned above is achieved in two ways. Firstly, a novel algorithm is developed to segment the nucleus and cytoplasm of white blood cell. Secondly, a model is built to extract the features and train the model. The different supervised classifiers are compared, and the one with the highest accuracy is used for the classification. Six hundred images are used in the experimentation. InfoGainAttributeEval and the Ranker Search method are used to achieve the feature selection which in turn helps in improvising the classifier performance. The result shows the classification of the acute lymphoblastic leukemia into its three respective categories namely: ALL-L1, ALL-L2, ALL-L3. The model can differentiate between a normal peripheral blood smear and an abnormal blood smear. The extracted feature values of a cancerous cell and a normal cell are also shown. The performance of the model is evaluated using the test images stained with various stains. The proposed algorithm achieved an overall accuracy of 98.6%. The promising results show that it can be used as a diagnostic tool by the pathologists.

Graphical abstract


















Similar content being viewed by others
References
Karen Seiter (2016) ALL types. In:Acute Lymphoblastic Leukemia Staging. Available via https://emedicine.medscape.com/article/2006661-overview. Accessed 16th Nov,2017
Afshar S, Abdolrahmani F, Tanha FV, Seif MZ, Taheri K (2011) Recognition and prediction of leukemia with artificial neural network (ANN). Med J Islam Repub Iran 25(1):35–39
Alomari YM, Azma RZ, Abdullah SNHS, Omar K (2014) Automatic detection and quantification of WBCs and RBCs using iterative structured circle detection algorithm. Computational and Mathematical Methods in Medicine 2014:1–14. https://doi.org/10.1155/2014/979302
Belekar SJ, Chougule SR (2015) WBC segmentation using morphological operation and SMMT operator-a review. International Journal of Innovative Research in Computer and Communication Engineering 3(1):434–440
Bhagvathi SL, Thomas NS (2016) An automatic system for detecting and counting RBC and WBC using fuzzy logic. ARPN-JEAS 11(11):6891–6894
Bhamare, Miss. Madhuri G. and D.S.Patil (2013) Automatic blood cell analysis by using digital image processing: a preliminary study .Int J Eng Res Technol 2(9):3137–3141.
Erosion operation (2017) https://in.mathworks.com/help/images/ref/imerode.html. Accessed 16th November, 2017
Joshi MD, Karode AH, Suralkar SR (2013) White blood cells segmentation and classification to detect acute leukemia. International journal of emerging trends Technology in Computer. Science 2(3):147–151
Marzuki NIC, Mahmood NH, Razak MAA (2015) Segmentation of white blood cell nucleus using active contour. J Teknol 74(6):115–118. https://doi.org/10.11113/jt.v74.4675
Mishra, S. J., & Deshmukh,A.P. (2014) Detection of Leukemia in Human Blood Sample based on Microscopic Images. International Journal of Advanced Research in Electronics and Communication Engineering, 1(3) :10-14.
Mulik V, Bhilare PM, Alhat S (2016) Analysis of acute lymphoblastic leukemia cells using digital image processing. International Journal for Scientific Research and Development 4(2):70–72
Nasir A, Mustafa N, Nasir NFM (2009) Application of thresholding technique in determining ratio of blood cells for leukemia detection. In Proceedings of the international conference on man-machine systems (ICoMMS 2009), 1–6
Putzu L, Ruberto CD (2013) White blood cells identification and counting from microscopic blood image. International journal of medical, health, biomedical, bioengineeringand Pharm Eng 7(1):20–27
Sadeghian F, Ramli AR, Seman Z, Khahar BHA, Saripan MI (2009) A Framework for White Blood Cell Segmentation in Microscopic Blood Images Using Digital Image Processing. Biological Procedures Online 11(1):196–206. https://doi.org/10.1007/s12575-009-9011-2
Thanh TTP, Pham GN, Park JH , Moon KS, Lee SH, Kwon KR (2017) Acute leukemia classification using convolution neural network in clinical decision support system. In Proc Of Computer Science & Information Technology, 49–53. https://doi.org/10.5121/csit.2017.71305
Vaghela HP, Pandya M, Modi H, Potdar MB (2015) Leukemia detection using digital image processing techniques. International Journal of Applied Information Systems 10(1):43–51
Wang M, Zhou X, Li F, Huckins J, King RW, Wong STC Novel cell segmentation and online learning algorithms for cell phase identification in automated time-lapse microscopy. In 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro 65–68. https://doi.org/10.1109/isbi.2007.356789
Wang Q, Wang J, Zhou M, Li Q, Wang Y (2017) Spectral-spatial feature-based neural network method for acute lymphoblastic leukemia cell identification via microscopic hyperspectral imaging technology. Biomedical Optics Express 8(6):3019–3028
Saeedizadeh Z, Dehnavi A, A.M., Rabbani H (2015) Automatic recognition of myeloma cells in microscopic images using bottleneck algorithm, modified watershed and SVM classifier. J Microsc 261(1):46–56. https://doi.org/10.1111/jmi.12314
Jiang K, Jiang QX, Xiong Y (2003) A novel white blood cell segmentation scheme using scale-space filtering and watershed clustering. Mach Learning Cybernetics 5:2820–2825
Kumar BR, Joseph DK, Sreenivas TV( 2002) Teager energy based blood cell segmentation. In proceedings of the 14th international conference on digital signal processing, Santorini, Greece, 619–622
Alreza ZKK, Karimian A (2016) A. Design a new algorithm to count white blood cells for classification leukemic blood image using machine vision system. In Proceedings of the 6th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 251–256
Reta C, Gonzalez JA, Diaz R, Guichard JS (2011) Leukocytes segmentation using Markov random fields. Adv Exp Med Biol 696:345–353
Madhloom HT, Kareem SA, Ariffin H (2011) An image processing application for the localization and segmentation of lymphoblast cell using peripheral blood images. J Med Syst 36:2149–2158
Houby EMFE (2018) Framework of computer aided diagnosis Systems for Cancer Classification Based on medical images. J Med Syst 42:157–167
Theera-Umpon N (2005) Patch-based white blood cell nucleus segmentation using fuzzy clustering. ECTI Transactions on Electrical Eng., Electronics, and Communications 3(1):15–19
Mohammed EA, Mohamed MMA, Naugler Christopher, Far BH (2013) Chronic lymphocytic leukemia cell segmentation from microscopic blood images using watershed algorithm and optimal thresholding. In Proc. of the 26th IEEE Can Con El Comp En(CCECE):1–5
Esti Suryani and Wiharto Wiharto and Nizomjon Polvonov (2015) Identification and Counting White Blood Cells and Red Blood Cells using Image Processing Case Study of Leukemia. International Journal of Computer Science & Network Solutions 2(6):35–49.
Khobragade S, Mor DD, Patil CY (2015) Detection of leukemia in microscopic white blood cell images. In Proc. of the 2015 international conference on information processing (ICIP) (ICIP):435–440. https://doi.org/10.1109/INFOP.2015.7489422
Cuevas E, Díaz M, Manzanares M, Zaldivar D, Perez-Cisnero M (2013) An improved computer vision method for white blood cells detection. Computational and Mathematical Methods in Medicine:1–14
Ahasan R, Ratul AU, Bakibillah ASM White Blood Cells Nucleus Segmentation from Microscopic Images of strained peripheral blood film during Leukemia and Normal Condition. In Proc. of the 2016 5th International Conference on Informatics, Electronics and Vision (ICIEV): 361–366. https://doi.org/10.1109/ICIEV.2016.7760026
Guo N, Zeng L, Wu Q (2007) A method based on multispectral imaging technique for white blood cell segmentation. Comput Biol Med 37:70–76
Sinha N, Ramakrishnan AG (2002) Blood cell segmentation using EM algorithm. In proc. Third Indian conference on computer vision. Graphics & Image Processing (ICVGIP):1–6.https://doi.org/10.1109/ICIEV.2016.7760026
Wang M, Chu R (2009) A novel white blood cell detection method based on boundary support vectors. In proc. of the 2009 IEEE international conference on systems. Man and Cybernetics:2595–2598
Nivaldo Medeiros(n.d.) Blood Smear Database. In: Atlas.Available via http://www.hematologyatlas.com/principalpage.htm. Accessed 16th November, 2017.http://www.hematologyatlas.com/principalpage.htm. Accessed 16 Nov 2017
Labati RD, Piuri V, Scotti F (2011) ALL -IDB: the acute lymphoblastic leukemia image database for image processing. In Proc. of the 2011 IEEE international conference on image processing 2045–2048. https://doi.org/10.1109/ICIP.2011.6115881
C.I.E primaries (n.d.) http://hyperphysics.phy-astr.gsu.edu/hbase/vision/cieprim.html. Accessed 16th November, 2017
Area open function (2015) https://in.mathworks.com/help/images/ref/bwareaopen.html. Accessed 16th November, 2017
Border clear (2015) https://in.mathworks.com/help/images/ref/imclearborder.html. Accessed 16th November, 2017
Watershed Transform (2013) https://blogs.mathworks.com/steve/2013/11/19/watershed-transform-question-from-tech-support/. Accessed: 16th November,2017
Gonzalez, R. C., R. E. Woods, and S. L. Eddins (2004) Digital Image Processing Using MATLAB. New Jersey, Pearson Prentice Hall
Measurement properties (2017) https://in.mathworks.com/help/images/ref/regionprops.html. Accessed: 16th November, 2017
Label the components (2015) https://in.mathworks.com/help/images/ref/bwlabel.html. Accessed: 16th November, 2017
Filling holes (2015) https://in.mathworks.com/help/images/ref/imfill.html. Accessed16th November, 2017
Extended maxima (2012) https://in.mathworks.com/help/images/ref/imextendedmax.html. Accessed 16th November, 2017
Impose minima (2003) https://in.mathworks.com/help/images/ref/imimposemin.html. Accessed: 16th November, 2017
Subrajeet Mohapatra (2013) Hematological image analysis for acute lymphoblastic leukemia detection and classification. Doctoral Dissertation, National Institute of Technology Rourkela
Histogram equalization (2015). Histeq function. In: Histogram Equalization. Available via https://in.mathworks.com/help/images/histogram-equalization.html . Accessed: 16th Nov, 2017 https://in.mathworks.com/help/images/histogram-equalization.html. Accessed: 16 Nov 2017
GLCM (2006) http://support.echoview.com/WebHelp/Windows_and_Dialog_Boxes/Dialog_Boxes/Variable_properties_dialog_box/Operator_pages/GLCM_Texture_Features.htm#About_the _GLCM_and_textures. Accessed 16th November, 2017
Almuallim H, Dietterich TG (1991) Learning with many irrelevant features. In: Proc. Ninth National conference on Artificial intelligence (AAAI-91), Anaheim, CA 2: 547–552
Kira K, Rendell LA. (1992) The feature selection problem: traditional methods and a newalgorithm. In: Proc. AAAI-92, San Jose, CA 122–126
MyHematalogy(2017) Leishman Stain . In : MyHematology. Available via https://myhematology.com/red-blood-cells/leishman-stain/ . Accessed: 16th Nov, 2017
Wright Stain Method Technical Data Sheet (2005) Wright Stain . In: Technical Data Sheets. Available via. https://www.emsdiasum.com/microscopy/technical/datasheet/26060.aspx. Accessed 16 Nov 2017
ALL-IDB (2011) https://homes.di.unimi.it/scotti/all/results.php. Accessed 16th January,2019
Attribute-Relation File Format (ARFF) (2008) ARFF. In: Attribute-Relation File Format (ARFF). Available via. https://www.cs.waikato.ac.nz/ml/weka/arff.html. Accessed 16 Nov 2017
Mohammed EA, Mohamed MM, Far BH, Naugler C (2014) Peripheral blood smear image analysis: a comprehensive review. Journal of Pathology Informatics 5(1):9. https://doi.org/10.4103/2153-3539.129442
Adjust operation (2017) https://in.mathworks.com/help/images/ref/imadjust.html. Accessed:16th November, 2017
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Acharya, V., Kumar, P. Detection of acute lymphoblastic leukemia using image segmentation and data mining algorithms. Med Biol Eng Comput 57, 1783–1811 (2019). https://doi.org/10.1007/s11517-019-01984-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-019-01984-1