Abstract
Glaucoma is a chronic disease that threatens eye health and can cause permanent blindness. Since there is no cure for glaucoma, early screening and detection are crucial for the prevention of glaucoma. Therefore, a novel method for automatic glaucoma screening that combines clinical measurement features with image-based features is proposed in this paper. To accurately extract clinical measurement features, an improved UNet++ neural network is proposed to segment the optic disc and optic cup based on region of interest (ROI) simultaneously. Some important clinical measurement features, such as optic cup to disc ratio, are extracted from the segmentation results. Then, the increasing field of view (IFOV) feature model is proposed to fully extract texture features, statistical features, and other hidden image-based features. Next, we select the best feature combination from all the features and use the adaptive synthetic sampling approach to alleviate the uneven distribution of training data. Finally, a gradient boosting decision tree (GBDT) classifier for glaucoma screening is trained. Experimental results based on the ORIGA dataset show that the proposed algorithm achieves excellent glaucoma screening performance with sensitivity of 0.894, accuracy of 0.843, and AUC of 0.901, which is superior to other existing methods.
Graphical abstract

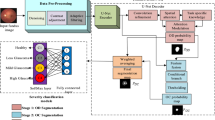
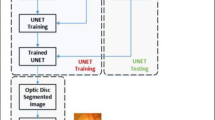
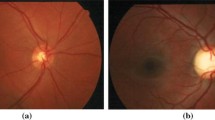
Framework of the proposed glaucoma classification method




















Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Tham Y-C, Li X, Wong TY, Quigley HA, Aung T, Cheng C-Y (2014) Global prevalence of glaucoma and projections of glaucoma burden through. Ophthalmology 121:2081–2090
Bourne RRA (2006) Worldwide glaucoma through the looking glass. Br J Ophthalmol 90:253–254
Shen SY, Wong TY, Foster PJ, Loo JL, Rosman M, Loon SC, Wong WL, Saw SM, Aung T (2008) The prevalence and types of glaucoma in Malay people: the Singapore Malay Eye Study. Invest Ophthalmol Vis Sci 49:3846–3851
Raghavendra U, Gudigar A, Bhandary SV, Raol TN, Ciaccio EJ, Acharya UR (2019) A two-layer sparse autoencoder for glaucoma identification with fundus images. J Med Syst 43:299
Zhao X, Guo F, Mai YX, Tang J, Duan XC, Zou BJ, Jiang LZ (2019) Glaucoma screening pipeline based on clinical measurements and hidden features. IET Image Process 13:2213–2223
Garway-Heath DF, Hitchings RA (1998) Quantitative evaluation of the optic nerve head in early glaucoma. Brit J Ophthalmol 82:352–361
Budenz DL, Barton K, Whiteside-de Vos J, Schiffman J, Bandi J, Nolan W, Herndon L, Kim H, Hay-Smith G, Tielsch JM (2014) Prevalence of glaucoma in an urban west african population. JAMA Ophthalmol 131:651–658
Derick R, Pasquale L, Pease M, Quigley H (1994) A clinical study of peripapillary crescents of the optic disc in chronic experimental glaucoma in monkey eyes. Arch Ophthalmol 112:846–850
Jonas JB, Bergua A, Schmitz-Valckenberg P et al (2000) Ranking of optic disc variables for detection of glaucomatous optic nerve damage. Invest Ophthalmol Vis Sci 41:1764–1773
Hancox OD, Michael D (1999) Optic disc size, an important consideration in the glaucoma evaluation. Clin Eye Vis Care 11:59–62
Yin F, Liu J, Wong DW, Tan NM et al (2012) Automated segmentation of optic cup and disc in fundus images for glaucoma diagnosis’, In: The 25th international symposium on computer-based medical systems (CBMS)
Smola A, Vishwanathan S (2008) Introduction to machine learning. Cambridge University Press
Nayak J, Acharya R, Bhat P, Shetty N, Lim T-C (2009) Automated diagnosis of glaucoma using digital fundus images. J Med Syst 33:337–346
Cheng J, Liu J, Xu Y, Yin F, Wong DWK, Tan NM, Tao D, Cheng CY, Aung T, Wong TY (2013) Superpixel classification based optic cup and disc segmentation for glaucoma screening. IEEE Trans Med Imaging 32:1019–1032
Noronhaam KP, Acharya UR, Nayaka KP, Martis RJ, Bhandary SV (2014) Automated classification of glaucoma stages using higher order cumulant features. Biomed Signal Proces Control 10:174–183
Acharyaa UR, Ng EYK, Eugenea LWJ, Noronha KP, Min LC, Nayak KP, Bhandarye SV (2015) Decision support system for the glaucoma using Gabor transformation. Biomed Signal Proces Control 15:18–26
Dua S, Acharya UR, Sree SV (2012) Wavelet-based energy features for glaucomatous image classifification. IEEE Trans Inf Technol Biomed 16:80–87
Haleem M, Han L, Hemert J, Fleming A, Pasquale LR, Silva PS, Song BJ, Aiello LP (2016) Regional image features model for automatic classification between normal and glaucoma in fundus and scanning laser ophthalmoscopy (SLO) images. J Med Syst 40:132
Long J, Shelhamer E, and Darrell T (2015) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intel 39:640 651
Sevastopolsky A Optic disc and cup segmentation methods for glaucoma detection with modification of U-Net convolutional neural. In: The 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Fu H, Cheng J, Xu Y, Wong DWK, Liu J, Cao XC (2018) Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans Med Imaging 37:1597–1605
Chen XY, Xu YW, Wong DWK, Wong WY, Liu J (2015) Glaucoma detection based on deep convolutional neural network. In: The 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
Diaz-Pinto A, Morales S, Naranjo V, Köhler T, Mossi JM, Navea A (2019) CNNs for automatic glaucoma assessment using fundus images: an extensive validation. Biomed Eng Online 18:29
Zhou ZW, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: a nested U-Net architecture for medical image segmentation. In: The 4th Deep Learning in Medical Image Analysis (DLMIA) Workshop
Lin T-Y, Goyal P, Girshick R, He KM, and Dollár P (2017) Focal loss-- focal loss for dense object detection. In: The IEEE International Conference on Computer Vision (ICCV)
He HB, Bai Y, Garcia EA, and Li S (2008) ADASYN: adaptive synthetic sampling approach for imbalanced learning. In: IEEE International Joint Conference on Neural Networks (IJCNN)
Ronneberger O, Fischer P, and Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: Proceedings of International Conference on Medical image computing and computer-assisted intervention
Huang G, Liu Z, Van Der Maaten L, and Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of CVPR
Vanajakshi B, Sujatha B, Srirama K (2010) A study on implementation of advanced morphological operations. IJCSNS Int J Comput Sci Netw Secur 10:6–9
Daugman JG (1988) Complete discrete 2-D Gabor transforms by neural networks forimage analysis and compression. IEEE Trans Acoust Speech Sig Process 36:1169–1179
Liu H, and Setiono R (1995) Chi2: Feature selection and discretization of numeric attributes. In: Proc. IEEE 7th Int. Conf. Tools with Artif. Intell
Kwak N, Choi C-H (2002) Input feature selection for classification problems. IEEE T Neural Netw 13:143–159
Vergara JR, Estevez PA (2014) A review of feature selection methods based on mutual information. Neural Comput Applic 24:175–186
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29:1189–1232
Rao H, Shi X, Rodrigue AK, Feng J, Xia Y, Elhoseny M, Yuan X, Gu L (2019) Feature selection based on artificial bee colony and gradient boosting decision tree. Appl Soft Comput 74:634–642
Zhang Z, Yin FS, Liu J, Wong WK (2010) Origa-light: An online retinal fundus image database for glaucoma analysis and research. In: Proceedings of Engineering in Medicine and Biology Society (EMBC), pp. 3065–3068
Almazroa A, Alodhayb S, Osman E et al (2018) Retinal fundus images for glaucoma analysis: the RIGA dataset. In: Proceeding of SPIE Medical Imaging. Houston, pp.1–8
Sivaswamy J, Krishnadas S, Chakravarty A et al (2015) A comprehensive retinal image dataset for the assessment of glaucoma from the optic nerve head analysis. JSM Biomed Imaging Data Papers 2:1–7
Decencière E, Zhang X, Cazuguel G, Lay B, Cochener B, Trone C, Gain P, Ordonez R, Massin P, Erginay A, Charton B, Klein JC (2014) Feedback on a publicly distributed image database: the Messidor database. Image Anal Stereol 33:231–234
Koh JE, Acharya UR, Hagiwara Y et al (2017) Diagnosis of retinal health in digital fundus images using continuous wavelet transform (CWT) and entropies. Comput Biol Med 84:89–97
Nugroho HA, Oktoeberza KZW, Adji TB, Najamuddin F (2015) Detection of exudates on color fundus images using texture-based feature extraction. Int J Technol 6:121
Funding
This work was supported by the National Natural Science Foundation of China under Grant 61502537, the Natural Science Foundation of Hunan Province of China under Grant 2018JJ3681, the Independent Exploration and Innovation Project for Graduate Students of Central South University under Grant 2020zzts567, and the Open Project Fund of the Key Laboratory of Digital Signal and Image Processing of Guangdong Province under Grant 2018GDDSIPL-01.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Guo, F., Li, W., Tang, J. et al. Automated glaucoma screening method based on image segmentation and feature extraction. Med Biol Eng Comput 58, 2567–2586 (2020). https://doi.org/10.1007/s11517-020-02237-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-020-02237-2
Keywords
Profiles
- Fan Guo View author profile