Abstract
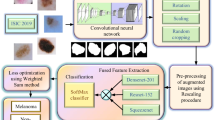
The automatic classification of skin lesions in dermoscopy images remains challenging due to the morphological diversity of skin lesions, the existence of intrinsic cutaneous features and artefacts, the lack of training data, and the insufficient recognition abilities of current methods. To meet these challenges, we construct a new densely connected convolutional network termed DenseSFNet-45, which is obtained by integrating our proposed novel architectural unit (an SE-Fire (SF) block) into the dense block of a dense convolutional network (DenseNet). The SF block consists of a cascade of a Fire module and a squeeze-and-excitation (SE) block, enhancing the representational power of DenseNet by exploiting both spatial and channel-wise information. Based on DenseSFNet, we propose a novel two-stage framework consisting of skin lesion segmentation followed by lesion classification to accurately classify skin lesions. The classification step is performed on the segmented lesion rather than the whole dermoscopy image, enabling the classification network to extract more specific and discriminative features. The proposed method is extensively evaluated on three public databases: ISBI 2017 Skin Lesion Analysis Towards Melanoma Detection Challenge dataset (ISBI-skin-2017), ISBI 2018 Skin Lesion Analysis Towards Melanoma Detection Challenge dataset (ISBI-skin-2018), and PH2 dataset. The experimental results demonstrate the superior performance of our method relative to that of the traditional machine learning algorithms, the existing classical classification models, baselines, and state-of-the-art methods.
Graphical abstract









Similar content being viewed by others
References
Jerant AF, Johnson JT, Sheridan CD, Caffrey TJ (2000) Early detection and treatment of skin cancer. American Family Physician 62(2):357–368
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA: A Cancer Journal for Clinicians 69(1):7–34
Freedberg KA, Geller AC, Miller DR, Lew RA, Koh HK (1999) Screening for malignant melanoma: a cost-effectiveness analysis. Journal of the American Academy of Dermatology 41(5):738–745
Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, Fleming ID, Gershenwald JE, Houghton A Jr, Kirkwood JM et al (2001) Final version of the American joint committee on cancer staging system for cutaneous melanoma. Journal of Clinical Oncology 19(16):3635–3648
Braun RP, Rabinovitz HS, Oliviero M, Kopf AW, Saurat JH (2005) Dermoscopy of pigmented skin lesions. Journal of the American Academy of Dermatology 52(1):109–121
Celebi ME, Kingravi HA, Uddin B, Iyatomi H, Aslandogan YA, Stoecker WV, Moss RH (2007) A methodological approach to the classification of dermoscopy images. Computerized Medical Imaging and Graphics 31(6):362–373
Schaefer G, Krawczyk B, Celebi ME, Iyatomi H (2014) An ensemble classification approach for melanoma diagnosis. Memetic Computing 6(4):233–240
Murugan A, Nair SAH, Kumar KS (2019) Detection of skin cancer using svm, random forest and knn classifiers. Journal of Medical Systems 43(8):1–9
Zaqout I (2019) Diagnosis of skin lesions based on dermoscopic images using image processing techniques. Pattern Recognition-Selected Methods and Applications
Almaraz-Damian JA, Ponomaryov V, Sadovnychiy S, Castillejos-Fernandez H (2020) Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual information measures. Entropy 22(4):484
Dhivyaa C, Sangeetha K, Balamurugan M, Amaran S, Vetriselvi T, Johnpaul P (2020) Skin lesion classification using decision trees and random forest algorithms. Journal of Ambient Intelligence and Humanized Computing pp 1–13
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proceedings of the IEEE 86(11):2278–2324
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Barata C, Celebi ME, Marques JS (2021) Explainable skin lesion diagnosis using taxonomies. Pattern Recognition 110:107413
Xie Y, Xia Y, Zhang J, Song Y, Feng D, Fulham M, Cai W (2018) Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest ct. IEEE Transactions on Medical Imaging 38(4):991–1004
Zhang J, Xie Y, Wu Q, Xia Y (2018) Skin lesion classification in dermoscopy images using synergic deep learning. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 12–20
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE transactions on pattern analysis and machine intelligence 40(4):834–848
He K, Gkioxari G, Dollár P, Girshick R (2017) Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision. pp 2961–2969
Lin TY, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision. pp 2980–2988
Yu Z, Jiang X, Zhou F, Qin J, Ni D, Chen S, Lei B, Wang T (2018) Melanoma recognition in dermoscopy images via aggregated deep convolutional features. IEEE Transactions on Biomedical Engineering 66(4):1006–1016
Mahbod A, Schaefer G, Ellinger I, Ecker R, Pitiot A, Wang C (2019) Fusing fine-tuned deep features for skin lesion classification. Computerized Medical Imaging and Graphics 71:19–29
Zhang J, Xie Y, Xia Y, Shen C (2019) Attention residual learning for skin lesion classification. IEEE Transactions on Medical Imaging 38(9):2092–2103
Hosny KM, Kassem MA, Fouad MM (2020) Classification of skin lesions into seven classes using transfer learning with alexnet. Journal of Digital Imaging 33(5):1325–1334
Narin A (2021) Accurate detection of covid-19 using deep features based on x-ray images and feature selection methods. Computers in Biology and Medicine 137:104771
Razzak I, Naz S (2020) Unit-vise: Deep shallow unit-vise residual neural networks with transition layer for expert level skin cancer classification. IEEE/ACM Transactions on Computational Biology and Bioinformatics
Yu L, Chen H, Dou Q, Qin J, Heng PA (2016) Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Transactions on Medical Imaging 36(4):994–1004
Al-Masni MA, Kim DH, Kim TS (2020) Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Computer Methods and Programs in Biomedicine 190:105351
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 4700–4708
Zhang K, Guo Y, Wang X, Yuan J, Ding Q (2019) Multiple feature reweight densenet for image classification. IEEE Access 7:9872–9880
Tao Y, Xu M, Lu Z, Zhong Y (2018) Densenet-based depth-width double reinforced deep learning neural network for high-resolution remote sensing image per-pixel classification. Remote Sensing 10(5):779
Liang S, Zhang R, Liang D, Song T, Ai T, Xia C, Xia L, Wang Y (2018) Multimodal 3d densenet for idh genotype prediction in gliomas. Genes 9(8):382
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K (2016) Squeezenet: Alexnet-level accuracy with 50x fewer parameters and \(<\) 0.5 mb model size. arXiv:1602.07360
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 7132–7141
Shan P, Wang Y, Fu C, Song W, Chen J (2020) Automatic skin lesion segmentation based on fc-dpn. Comput Biol Med 123:103762
Codella NC, Gutman D, Celebi ME, Helba B, Marchetti MA, Dusza SW, Kalloo A, Liopyris K, Mishra N, Kittler H, et al (2018) Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, pp 168–172
Tschandl P, Rosendahl C, Kittler H (2018) The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 5(1):1–9
Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, Helba B, Kalloo A, Liopyris K, Marchetti M, et al (2019) Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv:1902.03368
Mendonça T, Ferreira PM, Marques JS, Marcal AR, Rozeira J (2013) Ph 2-a dermoscopic image database for research and benchmarking. In: 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, pp 5437–5440
Lin M, Chen Q, Yan S (2013) Network in network. arXiv:1312.4400
Glorot X, Bordes A, Bengio Y (2011) Deep sparse rectifier neural networks. In: Proceedings of the fourteenth international conference on artificial intelligence and statistics. pp 315–323
Bottou L (2012) Stochastic gradient descent tricks. In: Neural networks: Tricks of the trade. Springer, pp 421–436
Gutman D, Codella NC, Celebi E, Helba B, Marchetti M, Mishra N, Halpern A (2016) Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (isbi) 2016, hosted by the international skin imaging collaboration (isic). arXiv:1605.01397
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems. pp 1097–1105
Woo S, Park J, Lee JY, Kweon IS (2018) Cbam: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV). pp 3–19
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017) Grad-cam: Visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision. pp 618–626
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Chollet F (2017) Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 1251–1258
Xie S, Girshick R, Dollár P, Tu Z, He K (2017) Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 1492–1500
Tan M, Le Q (2019) Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, PMLR, pp 6105–6114
Mahbod A, Schaefer G, Wang C, Ecker R, Ellinge I (2019) Skin lesion classification using hybrid deep neural networks. In: ICASSP 2019-2019 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE, pp 1229–1233
Harangi B (2018) Skin lesion classification with ensembles of deep convolutional neural networks. J Biomed Inform 86:25–32
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 61773068 and 61671141) and the Fundamental Research Funds for the Central Universities (No. N2224001-7).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shan, P., Fu, C., Dai, L. et al. Automatic skin lesion classification using a new densely connected convolutional network with an SF module. Med Biol Eng Comput 60, 2173–2188 (2022). https://doi.org/10.1007/s11517-022-02583-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02583-3