Abstract
Brain functional network (FN) has emerged as a potential tool for identifying mental and neurological diseases. Traditional FN estimation methods such as Pearson’s correlation (PC) and sparse representation (SR), despite their popularity, can only model low-order relationships between brain regions (i.e., nodes of FN), thus failing to capture more complex interaction in the brain. Recently, researchers proposed to estimate high-order FN (HoFN) and successfully used them in the early diagnosis of neurological diseases. In practice, however, such HoFN is constructed by directly considering the columns (or rows) of the adjacency matrix of low-order FN (LoFN) as node feature vectors that may contain some redundant or noisy information. In addition, it is not really reflected whether the original low-order relationship is maintained during the construction of the HoFN. To address these problems, we propose correlation-preserving embedding (COPE) to re-code the LoFN prior to constructing HoFN. Specifically, we first use SR to construct traditional LoFN. Then, we embed the LoFN via COPE to generate the new node representation for removing the potentially redundant/noisy information in original node feature vectors and simultaneously maintaining the low-order relationship between brain regions. Finally, the expected HoFN is estimated by SR based on the new node representation. To verify the effectiveness of the proposed scheme, we conduct experiments on 137 subjects from the public Alzheimer’s Disease Neuroimaging Initiative (ADNI) database to identify subjects with mild cognitive impairment (MCI) from normal controls. Experimental results show that the proposed scheme can achieve better performance than the baseline method.
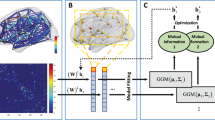
Graphical abstract
Brain functional network (FN) has emerged as a potential tool for identifying mental and neurological diseases. Traditional FN estimation methods such as Pearson’s correlation (PC) and sparse representation (SR), despite their popularity, can only model low-order relationships between brain regions (i.e., nodes of FN), thus failing to capture more complex interaction in the brain. Recently, researchers proposed to estimate high-order FN (HoFN) and successfully used them in the early diagnosis of neurological diseases. In practice, however, such HoFN is constructed by directly considering the columns (or rows) of the adjacency matrix of low-order FN (LoFN) as node feature vectors that may contain some redundant or noisy information. In addition, it is not really reflected whether the original low-order relationship is maintained during the construction of the HoFN. To address these problems, we propose correlation-preserving embedding (COPE) to re-code the LoFN prior to constructing HoFN. Specifically, we first use SR to construct traditional LoFN. Then, we embed the LoFN via COPE to generate the new node representation for removing the potentially redundant/noisy information in original node feature vectors and simultaneously maintaining the low-order relationship between brain regions. Finally, the expected HoFN is estimated by SR based on the new node representation. To verify the effectiveness of the proposed scheme, we conduct experiments on 137 subjects from the public Alzheimer’s Disease Neuroimaging Initiative (ADNI) database to identify subjects with mild cognitive impairment (MCI) from normal controls. Experimental results show that the proposed scheme can achieve better performance than the baseline method.









Similar content being viewed by others
References
Liu Y et al (2008) Regional homogeneity, functional connectivity and imaging markers of Alzheimer’s disease: a review of resting-state fMRI studies. Neuropsychologia 46(6):1648–1656
Azeez AK, Biswal BB (2017) A review of resting-state analysis methods. Neuroimaging Clin 27(4):581–592
Geschwind DH, Levitt P (2007) Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol 17(1):103–111
Simonoff E, Pickles A, Charman T, Chandler S, Loucas T, Baird G (2008) Psychiatric disorders in children with autism spectrum disorders: prevalence, comorbidity, and associated factors in a population-derived sample. J Am Acad Child Adolesc Psychiatry 47(8):921–929
Greicius MD et al (2007) Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiat 62(5):429–437
Cullen KR et al (2014) Abnormal amygdala resting-state functional connectivity in adolescent depression. JAMA Psychiat 71(10):1138–1147
Wang L et al (2006) Changes in hippocampal connectivity in the early stages of Alzheimer’s disease: evidence from resting state fMRI. Neuroimage 31(2):496–504
Rombouts SA, Barkhof F, Goekoop R, Stam CJ, Scheltens P (2005) Altered resting state networks in mild cognitive impairment and mild Alzheimer’s disease: an fMRI study. Hum Brain Mapp 26(4):231–239
Reijneveld JC, Ponten SC, Berendse HW, Stam CJ (2007) The application of graph theoretical analysis to complex networks in the brain. Clin Neurophysiol 118(11):2317–2331
Wang J, Zuo X, He Y (2010) Graph-based network analysis of resting-state functional MRI. Front Syst Neurosci 4:16
Pervaiz U, Vidaurre D, Woolrich MW, Smith SM (2020) Optimising network modelling methods for fMRI. Neuroimage 211:116604
Qiao L, Zhang H, Kim M, Teng S, Zhang L, Shen D (2016) Estimating functional brain networks by incorporating a modularity prior. Neuroimage 141:399–407
Friston KJ, Harrison L, Penny W (2003) Dynamic causal modelling. Neuroimage 19(4):1273–1302
Smith SM et al (2013) Functional connectomics from resting-state fMRI. Trends Cogn Sci 17(12):666–682
Schwarz AJ, McGonigle J (2011) Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. Neuroimage 55(3):1132–1146
van den Heuvel MP, de Lange SC, Zalesky A, Seguin C, Yeo BT, Schmidt R (2017) Proportional thresholding in resting-state fMRI functional connectivity networks and consequences for patient-control connectome studies: Issues and recommendations. Neuroimage 152:437–449
Huang S et al (2010) Learning brain connectivity of Alzheimer’s disease by sparse inverse covariance estimation. Neuroimage 50(3):935–949
Tan Z, Yang P, Nehorai A (2013) Joint-sparse recovery in compressed sensing with dictionary mismatch. 2013 5th IEEE International Workshop on Computational Advances in Multi-Sensor Adaptive Processing (CAMSAP) 248–251. https://doi.org/10.1109/CAMSAP.2013.6714054
Plis SM et al (2014) High-order interactions observed in multi-task intrinsic networks are dominant indicators of aberrant brain function in schizophrenia. Neuroimage 102:35–48
Macke JH, Opper M, Bethge M (2011) Common input explains higher-order correlations and entropy in a simple model of neural population activity. Phys Rev Lett 106(20):208102
Guo H, Liu L, Chen J, Xu Y, Jie X (2017) Alzheimer classification using a minimum spanning tree of high-order functional network on fMRI dataset. Front Neurosci 11:639
Zhou Y, Qiao L, Li W, Zhang L, Shen D (2018) Simultaneous estimation of low-and high-order functional connectivity for identifying mild cognitive impairment. Front Neuroinform 12:3
Chen X, Zhang H, Lee S-W, Shen D (2017) Hierarchical high-order functional connectivity networks and selective feature fusion for MCI classification. Neuroinformatics 15(3):271–284
Zhao F, Zhang H, Rekik I, An Z, Shen D (2018) Diagnosis of autism spectrum disorders using multi-level high-order functional networks derived from resting-state functional mri. Front Hum Neurosci 12:184
Chen X et al (2016) High-order resting-state functional connectivity network for MCI classification. Hum Brain Mapp 37(9):3282–3296
Zhang H et al (2016) Topographical information-based high-order functional connectivity and its application in abnormality detection for mild cognitive impairment. J Alzheimer’s Dis 54(3):1095–1112
Jia X, Zhang H, Adeli E, Shen D (2017) Consciousness level and recovery outcome prediction using high-order brain functional connectivity network. International Workshop on Connectomics in Neuroimaging 17–24. First online: 02 Sept 2017
Guo T, Zhang Y, Xue Y, Qiao L, Shen D (2021) Brain function network: higher order vs. more discrimination. Front Neurosci 1033. https://doi.org/10.3389/fnins.2021.696639
Zhou Y, Zhang L, Teng S, Qiao L, Shen D (2018) Improving sparsity and modularity of high-order functional connectivity networks for MCI and ASD identification. Front Neurosci 12:959
Sun L, Xue Y, Zhang Y, Qiao L, Zhang L, Liu M (2021) Estimating sparse functional connectivity networks via hyperparameter-free learning model. Artif Intell Med 111:102004
Tzourio-Mazoyer N et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15(1):273–289
Li W, Wang Z, Zhang L, Qiao L, Shen D (2017) Remodeling Pearson’s correlation for functional brain network estimation and autism spectrum disorder identification. Front Neuroinform 11:55
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol (TIST) 2(3):1–27
Dadi K et al (2019) Benchmarking functional connectome-based predictive models for resting-state fMRI. Neuroimage 192:115–134
Guyon I, Elisseeff A (2003) An introduction to variable and feature selection. J Mach Learn Res 3(Mar):1157–1182
Gómez S, Jensen P, Arenas A (2009) Analysis of community structure in networks of correlated data. Phys Rev E 80(1):016114
Rubinov M, Sporns O (2011) Weight-conserving characterization of complex functional brain networks. Neuroimage 56(4):2068–2079
Jiang X, Zhang L, Qiao L, Shen D (2019) Estimating functional connectivity networks via low-rank tensor approximation with applications to MCI identification. IEEE Trans Biomed Eng 67(7):1912–1920
Xue Y, Zhang L, Qiao L, Shen D (2020) Estimating sparse functional brain networks with spatial constraints for MCI identification. PLoS ONE 15(7):e0235039
Greicius M (2008) Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol 21(4):424–430
Albert MS et al (2011) The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement 7(3):270–279
Granger CW (1969) Investigating causal relations by econometric models and cross-spectral methods. Econometrica: J Econ Soc 37:424–438
Mclntosh A, Gonzalez-Lima F (1994) Structural equation modeling and its application to network analysis in functional brain imaging. Hum Brain Mapp 2(1–2):2–22
Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C (2010) Six problems for causal inference from fMRI. Neuroimage 49(2):1545–1558
Smith SM et al (2011) Network modelling methods for FMRI. Neuroimage 54(2):875–891
Theiler J, Eubank S, Longtin A, Galdrikian B, Farmer JD (1992) Testing for nonlinearity in time series: the method of surrogate data. Physica D: Nonlinear Phenom 58(1–4):77–94
Steuer R, Kurths J, Daub CO, Weise J, Selbig J (2002) The mutual information: detecting and evaluating dependencies between variables. Bioinformatics 18(suppl_2):S231–S240
Hamilton WL (2020) Graph representation learning. Synth Lect Artif Intell Mach Learn 14(3):1–159
Funding
This work was partly supported by the National Natural Science Foundation of China (Nos. 61976110, 62176112, 11931008), the Natural Science Foundation of Shandong Province (No. ZR202102270451), and the Open Project of Liaocheng University Animal Husbandry Discipline (No. 319312101–01).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Su, H., Zhang, L., Qiao, L. et al. Estimating high-order brain functional networks by correlation-preserving embedding. Med Biol Eng Comput 60, 2813–2823 (2022). https://doi.org/10.1007/s11517-022-02628-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-022-02628-7