Abstract
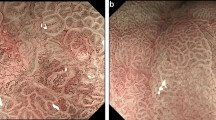
Esophageal squamous cell carcinoma (ESCC) is one of the most common histological types of esophageal cancers. It can seriously affect public health, particularly in Eastern Asia. Early diagnosis and effective therapy of ESCC can significantly help improve patient prognoses. The visualization of intrapapillary capillary loops (IPCLs) under magnification endoscopy (ME) can greatly support the identification of ESCC occurrences by endoscopists. This paper proposes an artificial-intelligence-assisted endoscopic diagnosis approach using deep learning for localizing and identifying IPCLs to diagnose early-stage ESCC. An improved Faster region-based convolutional network (R-CNN) with a polarized self-attention (PSA)-HRNetV2p backbone was employed to automatically detect IPCLs in ME images. In our study, 2887 ME with blue laser imaging (ME-BLI) images of 246 patients and 493 ME with narrow-band imaging (ME-NBI) images of 81 patients were collected from multiple hospitals and used to train and test our detection model. The ME-NBI images were used as the external testing set to verify the generalizability of the model. The experimental evaluation revealed that the proposed method achieved a recall of 79.25%, precision of 75.54%, F1-score of 0.764 and mean average precision (mAP) of 74.95%. Our method outperformed other existing approaches in our evaluation. It can effectively improve the accuracy of ESCC detection and provide a useful adjunct to the assessment of early-stage ESCC for endoscopists.













Similar content being viewed by others
References
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin D, Piñeros M, Znaor A, Bray F (2019) Estimating the global cancer incidence and mortality in: 2018 Globocan sources and methods. Int J Cancer 144(8):1941–1953
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin 68(6):394–424
Naveed M, Kubiliun N (2018) Endoscopic treatment of early-stage esophageal cancer. Curr Oncol Rep 20(9):71
Kuwano H, Nishimura Y, Oyama T, Kato H, Kitagawa Y, Kusano M, Shimada H, Takiuchi H, Toh Y, Doki Y, Naomoto Y, Matsubara H, Miyazaki T, Muto M, Yanagisawa A (2015) Guidelines for diagnosis and treatment of carcinoma of the esophagus April 2012 edited by the Japan esophageal society. Esophagus 12(1):1–30
Yada T, Yokoi C, Uemura N (2013) The current state of diagnosis and treatment for early gastric cancer. Diagnostic and therapeutic endoscopy
Shinozaki S, Osawa H, Hayashi Y, Lefor AK, Yamamoto H (2019) Linked color imaging for the detection of early gastrointestinal neoplasms. Ther Adv Gastroenterol 12
Diao W, Huang X, Shen L, Zeng Z (2018) Diagnostic ability of blue laser imaging combined with magnifying endoscopy for early esophageal cancer. Dig Liver Dis 50(10):1035–1040
Goda K, Tajiri H, Ikegami M, Yoshida Y, Yoshimura N, Kato M, Sumiyama K, Imazu H, Matsuda K, Kaise M, Kato T, Omar S (2009) Magnifying endoscopy with narrow band imaging for predicting the invasion depth of superficial esophageal squamous cell carcinoma. Dis Esophagus 22 (5):453–460
Oyama T, Inoue H, Arima M, Momma K, Omori T, Ishihara R, Hirasawa D, Takeuchi M, Tomori A, Goda K (2017) Prediction of the invasion depth of superficial squamous cell carcinoma based on microvessel morphology: magnifying endoscopic classification of the Japan esophageal society. Esophagus 14(2):105–112
Goda K, Irisawa A (2020) Japan esophageal society classification for predicting the invasion depth of superficial esophageal squamous cell carcinoma: Should it be modified now? Dig Endosc 32(1):37–38
Syed T, Doshi A, Guleria S, Syed S, Shah T (2020) Artificial intelligence and its role in identifying esophageal neoplasia. Dig Dis Sci 65(12):3448–3455
Zhang YH, Guo LJ, Yuan XL, Hu B (2020) Artificial intelligence-assisted esophageal cancer management: Now and future. World J Gastroenterol 26(35):5256–5271. https://doi.org/10.3748/wjg.v26.i35.5256
Lazăr DC, Avram MF, Faur AC, Goldiş A, Romoşan I, Tăban S, Cornianu M (2020) The impact of artificial intelligence in the endoscopic assessment of premalignant and malignant esophageal lesions: Present and future. Medicina 56(7)
Wang J, Sun K, Cheng T, Jiang B, Deng C, Zhao Y, Liu D, Mu Y, Tan M, Wang X, Liu W, Xiao B (2021) Deep high-resolution representation learning for visual recognition. IEEE Trans Pattern Anal Mach Intell 43(10):3349–3364. https://doi.org/10.1109/TPAMI.2020.2983686
Ren S, He K, Girshick R, Sun J (2015) Faster R-CNN: Towards real-time object detection with region proposal networks. Advances in neural information processing systems 28
Sun K, Zhao Y, Jiang B, Cheng T, Xiao B, Liu D, Mu Y, Wang X, Liu W, Wang J (2019) High-resolution representations for labeling pixels and regions. arXiv:190404514
Lin TY, Dollár P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2117–2125
Liu H, Liu F, Fan X, Huang D (2021) Polarized self-attention: Towards high-quality pixel-wise regression. arXiv:210700782
Zhang C, Ma L, Matsuura N, Tam P, Teoh A (2017) Tu1217 the use of convolutional neural artificial intelligence network to aid the diagnosis and classification of early esophageal neoplasia. A feasibility study. Gastroint Endosc 85:AB581–AB582
Xue DX, Zhang R, Feng H, Wang YL (2016) CNN-SVM for microvascular morphological type recognition with data augmentation. J Med Biol Eng 36(6):755–764
Zhao Y, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM (2019) Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy 51(4):333–341
Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T (2020) Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc 91(2):301–309.e1
Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T (2019) Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc 90(3):407–414
Everson M, Herrera L, Li W, Luengo I, Ahmad OF, Banks M, Magee CG, Alzoubaidi D, Hsu HM, Graham D, Vercauteren TKM, Lovat LB, Ourselin S, Kashin S, Wang H, Wang WL, Haidry RJ (2019) Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United Eur Gastroenterol J 7(2):297–306
García-Peraza-Herrera LC, Everson M, Lovat LB, Wang H, Wang WL, Haidry RJ, Stoyanov D, Ourselin S, Vercauteren TKM (2020) Intrapapillary capillary loop classification in magnification endoscopy: open dataset and baseline methodology. Int J Comput Assist Radiol Surg 15(4):651–659
Jie Guo L, Xiao X, Wu C, Zeng X, Hang Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B (2019) Real-time automated diagnosis of precancerous lesion and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc 91(1):41–51
Bochkovskiy A, Wang CY, Liao HYM (2020) YOLOV4: Optimal speed and accuracy of object detection. arXiv:200410934
Frey BJ, Dueck D (2007) Clustering by passing messages between data points. Science 315 (5814):972–976
Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering. In: International conference on medical image computing and computer-assisted intervention, pp 130–137
Zhao ZQ, Zheng P, Xu ST, Wu X (2019) Object detection with deep learning : A review. IEEE Trans Neural Netw Learn Syst 30(11):3212–3232. https://doi.org/10.1109/TNNLS.2018.2876865
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:14091556
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Sun K, Xiao B, Liu D, Wang J (2019) Deep high-resolution representation learning for human pose estimation. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 5693–5703
Solovyev R, Wang W, Gabruseva T (2021) Weighted boxes fusion: Ensembling boxes from different object detection models. Image Vision Comput 107:104,117
Neubeck A, Van Gool L (2006) Efficient non-maximum suppression. In: 18th international conference on pattern recognition (ICPR’06), vol 3, pp 850–855, DOI https://doi.org/10.1109/ICPR.2006.479
Bodla N, Singh B, Chellappa R, Davis LS (2017) Soft-NMS–improving object detection with one line of code. In: Proceedings of the IEEE international conference on computer vision, pp 5561–5569. https://doi.org/10.1109/ICCV.2017.593
Redmon J, Farhadi A (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7263–7271
Everingham M, Van Gool L, Williams CK, Winn J, Zisserman A (2010) The pascal visual object classes (VOC) challenge. Int J Comput Vision 88(2):303–338
Liu W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu CY, Berg AC (2016) SSD: Single shot multibox detector. In: European conference on computer vision, pp 21–37
Lin TY, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, pp 2999–3007. https://doi.org/10.1109/ICCV.2017.324
Zhang H, Wang Y, Dayoub F, Sunderhauf N (2021) Varifocalnet: An iou-aware dense object detector. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 8514–8523
Pang J, Chen K, Shi J, Feng H, Ouyang W, Lin D (2019) Libra R-CNN: Towards balanced learning for object detection. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 821–830
Liu S, Qi L, Qin H, Shi J, Jia J (2018) Path aggregation network for instance segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 8759–8768
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Zhang H, Zu K, Lu J, Zou Y, Meng D (2021) EPSANet: An efficient pyramid squeeze attention block on convolutional neural network. arXiv:210514447
Woo S, Park J, Lee JY, Kweon IS (2018) CBAM: Convolutional block attention module. In: Proceedings of the European conference on computer vision (ECCV), pp 3–19
Acknowledgements
Thanks to Department of the Gastroenterology, Zhongda Hospital Affiliated to Southeast University, for providing endoscopic images. All images were de-identified prior to their transfer to the study’s investigators, and the design and execution of this study complied with regional regulations and was approved by the Institutional Review Board (IRB) of Zhongda Hospital Affiliated to Southeast University.
Funding
This research was partially funded by the Jiangsu Provincial Key Research and Development Program under the grant number BE2019710, the Suzhou Municipal Science and Technology Bureau under the grant number SYS2019008 and the Changzhou Municipal Science and Technology Program under the grant number CE20195001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Long, Q., Liang, Y. et al. AI-assisted identification of intrapapillary capillary loops in magnification endoscopy for diagnosing early-stage esophageal squamous cell carcinoma: a preliminary study. Med Biol Eng Comput 61, 1631–1648 (2023). https://doi.org/10.1007/s11517-023-02777-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02777-3