Abstract
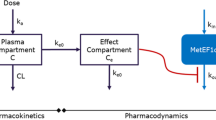
A new therapeutic approach against cancer is developed by the firm Erytech. This approach is based on starved cancer cells of an amino acid essential to their growth (the L-methionine). The depletion of plasma methionine level can be induced by an enzyme, the methionine-γ-lyase. The new therapeutic formulation is a suspension of erythrocytes encapsulating the activated enzyme. Our work reproduces a preclinical trial of a new anti-cancer drug with a mathematical model and numerical simulations in order to replace animal experiments and to have a deeper insight on the underlying processes. With a combination of a pharmacokinetic/pharmacodynamic model for the enzyme, substrate, and co-factor with a hybrid model for tumor, we develop a “global model” that can be calibrated to simulate different human cancer cell lines. The hybrid model includes a system of ordinary differential equations for the intracellular concentrations, partial differential equations for the concentrations of nutrients and drugs in the extracellular matrix, and individual based model for cancer cells. This model describes cell motion, division, differentiation, and death determined by the intracellular concentrations. The models are developed on the basis of experiments in mice carried out by Erytech. Parameters of the pharmacokinetics model were determined by fitting a part of experimental data on the concentration of methionine in blood. Remaining experimental protocols effectuated by Erytech were used to validate the model. The validated PK model allowed the investigation of pharmacodynamics of cell populations. Numerical simulations with the global model show cell synchronization and proliferation arrest due to treatment similar to the available experiments. Thus, computer modeling confirms a possible effect of treatment based on the decrease of methionine concentration. The main goal of the study is the development of an integrated pharmacokinetic/pharmacodynamic model for encapsulated methioninase and of a mathematical model of tumor growth/regression in order to determine the kinetics of L-methionine depletion after co-administration of Erymet product and Pyridoxine.











Similar content being viewed by others
Notes
\(T= \frac {tln(2)}{ln(C_{t})-ln(C_{0})}\), with T the doubling time, t the duration of the culture, and Ct and C0 respectively the cell concentration at time 0 and t.
Homocysteine is remethyled to methionine by transfer of a methyl group from N-5-methyltetrahydrofolate (CH3 − H4folate) calalyzed by methionine synthase (N-5-methyltetrahydrofolate) an enzyme that requires Vitamin B12 as a co-factor [28, 30].
$$CH_{3}-H_{4}folate+ Homocysteine \rightarrow H_{4}-folate+Methionine$$Homocysteine is the substrate of the B6 dependent enzyme cyasthionine β synthase which catalyzes an irreversible reaction (under physiological conditions) degrading Homocysteine into Cystasthiomine and Cysteine [28, 29].
$$ \begin{array}{@{}rcl@{}} Homocysteine + serine &\rightarrow& Cystathiomine + h_{2} O\\ Homocysteine + cysteine &\rightarrow& Cystathiomine + h_{2} S \end{array} $$This work participates to a marketing authorization application for a drug. In French, AMM: autorisation de mise sur le marché.
References
Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM (1983) The metabolic defect of methionine occurs frequently in human tumor cell lines. Biochem Biophys Res Commun 117(2)
Judde JG, Frost P (1988) Patterns of methionine auxotrophy in normal and neoplastic cells: the methionine independence of lymphocyte mitogenesis and low frequency of the methionine-dependent phenotype in human. Tumors Cancer Res 48:6775–6779
Kokkinakis DM, Schold, SC Jr, Hori H, Nobori T (1997) Effect of long-term depletion of plasma methionine on the growth and survival of human brain tumor xenografts in athymic mice. Nutr Cancer 29(3):195–204. https://doi.org/10.1080/01635589709514624
Hoshiya Y, Guo H, Kubota T, Inada T, Asanuma F, Yamada Y, Koh J, Kitajima M, Hoffman RM (1995) Human tumors are methionine-dependent in vivo. Anticancer Res 15:717–718
Pavillard V, Drbal AAA, Swaine DJ, Phillips RM, Double JA, Nicolaou A (2004) Analysis of cell-cycle kinetics and sulfur amino acid metabolism in methionine-dependent tumor cell lines; the effect of homocysteine supplementation. Biochem Pharmacol 67:1587–1599
Tan Y, Zavala, J Sr, Xu M, Zavala, J Jr, Hoffman RM (1996) Serum methionine depletion without side effects by methioninase in metastatic breast cancer patients. Anticancer Res 16(6C):3937–42
Tan Y, Zavala, J Sr, Han Q, Xu M, Sun X, Tan X, Magana R, Geller J, Hoffman RM (1997) Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res 17(5B):3857–60
Gilbert H (1999) Basic concepts in Biochemistry. ISBN: 0071356576
Gérard C, Goldbeter A (2015) Dynamics of the mammalian cell cycle in physiological and pathological conditions. WIREs Syst Biol Med. https://doi.org/10.1002/wsbm.1325
Hens JR, Sinha I, Perodin F, Cooper T, Sinha R, Plummer J, Perrone CE, Orentreich D (2016) Methionine-restricted diet inhibits growth of MCF10AT1-derived mammary tumors by increasing cell cycle inhibitors in athymic nude mice. Hens othersBMC Cancer 16:349. https://doi.org/10.1186/s12885-016-2367-1
Dua P, Hawkins E, van der Graaf PH (2015) A tutorial on target-mediated drug disposition (TMDD) models. CPTPharmacometricsSyst Pharmacol 4:324–33
Fischer S, Kurbatova P, Bessonov N, Gandrillon O, Volpert V, Crauste F (2012) Modelling erythroblastic islands: using a hybrid model to assess the function of central macrophage. J Theo Biol 298:92–106
Deutsch A (2007) Lattice-gas cellular automaton modeling of developing cell systems. Single-Cell-Based Models in Biology and Medicine, Mathematics and Biosciences in Interaction, 29–51
Dallon JC (2007) Models with lattice-free center-based cells interacting with continuum environment variables. Single Cell Based Models in Biology and Medicine, Mathematics and Biosciences in Interaction, III, 197–219
Eymard N, Bessonov N, Gandrillon O, Koury MJ, Volpert V (2015) The role of spatial organization of cells in erythropoiesis. J Math Biol 70(1):71–97
Alarcón T, Byrneb HM, Mainia PKA (2003) cellular automaton model for tumour growth in inhomogeneous environment. J Theor Biol 225:257–274
Anderson ARA, Chaplain MAJ (1998) Continuous discrete mathematical models of tumor-induced angiogenesis. Bull Math Biol 60(5):857–99
Anderson ARA, Chaplain MAJ, Rejniak KA (2007) The cellular Potts model and its variants. Single-Cell-Based Models in Biology and Medicine
Kurbatova P, Eymard N, Volpert V (2013) Hybrid model of erythropoiesis. Acta Biotheor 61(3):305–315
Eymard N, Kurbatova P (2015) Hybrid models in hematopoiesis. Math Model Nat Phenom 10(1):48–63
Bessonov N, Crauste F, Fischer S, Kurbatova P, Volpert V (2011) Application of hybrid models to blood cell production in the bone marrow. Math Model Nat Phenom 6(7):2–12
Bessonov N, Kurbatova P, Volpert V (2010) Particle dynamics modelling of cell populations. Math Model Nat Phenom 5(7):42–47. JANO9 The 9th International Conference on Numerical Analysis and Optimization
Kurbatova P, Bernard S, Bessonov N, Crauste F, Demin I, Dumontet C, Fischer S, Volpert V (2011) Hybrid model of erythropoiesis and leukemia treatment with cytosine arabinoside. SIAM J App Math 71(6):2246–2268
Sun X, Yang Z, Li S, Tan Y, Zhang N, Wang X, Yagi S, Yoshioka T, Takimoto A, Mitsushima K, Suginaka A, Frenkel EP, Hoffman RM (2003) In vivo efficacy of recombinant methioninase is enhanced by the combination of polyethylene glycol conjugation and pyridoxal 5-phosphate supplementation. Cancer Res 63:8377–8383
Yang Z, Wang J, Yoshioka T, Li B, Lu Q, Li S, Sun X, Tan Y, Yagi S, Frenkel EP, Robert M, Hoffman RM (2004) Pharmacokinetics methionine depletion, and antigenicity of recombinant methioninase in primates. Clin Cancer Res 10:2131–2138
Hoffman RM, Erbe RW (1976) High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci USA 73:1523–1527
Blom HJ, Smulders Y (2011) Overview of homocysteine and folate metabolism. With special references to cardiovascular disease and neural tube defects. J Inherit Metab Dis 34:75–81. https://doi.org/10.1007/s10545-010-9177-4
Medina MA, Urdiales JL, Amores-Sanchez MI (2001) Roles of homocysteine in cell metabolism Old and new functions. Eur J Biochem 268:3871–3882
Stipanuk MH, Ueki I (2011) Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur. J Inherit Metab Dis 34(1):17–32. https://doi.org/10.1007/s10545-009-9006-9
Banerjee RV, Matthews RG (1990) Cobalamin-dependent methionine synthase. FASEB J 4 (5):1450–9
Poomipark N, Flatley JE, Hill MH, Mangnall B, Azar E, Grabowski P, Powers HJ (2016) Methyl donor status influences DNMT expression and Global DNA methylation in cervical cancer cells. Asian Pac J Cancer Prev :17
Hu J, Cheung N-KV (2009) Methionine depletion with recombinant methioninase: In vitro and in vivo efficacy against neuroblastoma and its synergism with chemotherapeutic drugs. Int J Cancer 124(7):1700–1706. https://doi.org/10.1002/ijc.24104
Hoffman RM (2017) Is DNA methylation the new guardian of the genome? Mol Cytogenet 10:11. https://doi.org/10.1186/s13039-017-0314-8. eCollection 2017
Murakami T, Li S, Han Q, Tan Y, Kiyuna T, Igarashi K, Kawaguchi K, Hwang HK, Miyake K, Singh AS, Nelson SD, Dry SM, Li Y, Hiroshima Y, Lwin TM, DeLong JC, Chishima T, Tanaka K, Bouvet M, Endo I, Eilber FC, Hoffman RM (2017) Recombinant methioninase effectively targets a Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget. https://doi.org/10.18632/oncotarget.15823.
Lu S, Epner DE (2000) Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr Cancer 38(1):123–130
Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P (2003) Methionine dependency and cancer treatment. Cancer Treat Rev 29:489–499. https://doi.org/10.1016/S0305-7372(03)00118-X
El-Sayed ASA, Ruff LE, Ghany SEA, Ali GS, Esener S (2016) Molecular and spectroscopic characterization of aspergillus flavipes and pseudomonas putida L-Methionine γ-Lyase in Vitro. Appl Biochem Biotechnol https://doi.org/10.1007/s12010-016-2299-x
Özkan YE, Yardim-Akaydin S, Firat H, Çalişkan-Can E, Ardiç S, Şimşek B (2007) Usefulness of homocysteine as a cancer marker: Total thiol compounds and folate levels in untreated lung cancer patients. Anticancer Res 27:1185–1190
Fischer S, Kurbatova P, Bessonov N, Gandrillon O, Volpert V, Crauste F (2012) Modelling erythroblastic islands: using a hybrid model to assess the function of central macrophage. Theo Biol 298:92–106
Bessonov N, Demin I, Pujo-Menjouet L, Volpert V (2009) A multi-agent model describing self-renewal of differentiation effects on the blood cell population. Math Comput Model :2116–2127
Bessonov N, Crauste F, Fischer S, Kurbatova P, Volpert V (2011) Application of hybrid models to blood cell production in the bone marrow. Math Model Nat Phenom (7):2–12
Kurbatova P, Eymard N, Volpert V (2013) Hybrid model of erythropoiesis. Acta Biotheor 61(3):305–315
Eymard N, Kurbatova P (2015) Hybrid models in hematopoiesis and in Megakaryopoiesis. Math Model Nat Phenom 10(1):48–63
Eymard N, Bessonov N, Gandrillon O, Koury MJ, Volpert V (2015) The role of spatial organization of cells in erythropoiesis. J Math Biol 70(1):71–97
(2011) Hybrid modelling of erythropoiesis and blood disorders. Polina Kurbatova Thesis
Dayneka NL, Garg V, Jusko WJ (1993) Comparison of four basic models of indirect pharmacodynamic responses. J Pharmacokinet Biopharm 21(4):457–478
Goutelle S, Maurin M, Rougier F, Barbaut X, Bourguignon L, Ducher M, Maire P (2008) The Hill equation: a review of its capabilities in pharmacological modelling. Soc Française Pharmacol de Thérapeutique Fundam Clin Pharmacol 22:633–648
Bellu G, Saccomani MP, Audoly S, D’Angio L (2007) DAISY: A new software tool to test global identifiability of biological and physiological systems. Comput Methods Programs Biomed 88(1):52–61
Gross E, Meshkat N, Shiu A Identifiability of linear compartmental models: The singular locus. Available online 14 October 2021, Version of Record 15 November 2021
Godfrey KR, Jones RP, Brown RF The identifiability of linear models
Model Identifiability. Paola Lecca, Chapter of the book “Identifiability and Regression Analysis of Biological Systems Models” First Online: 06 March 2020
Acknowledgements
We thank Erytech pharma for its contribution to this work. We particularly thank doctors Vanessa Bourgeaux and Karine Sénéchal for the collected data. The work has been supported by the RUDN University Strategic Academic Leadership Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This work was funded by Erytech pharma. N. Eymard and P. Kurbatova were employed by EZUS, private subsidiary of Lyon 1 university. F. Gueyffier and P. Nony declared no conflict of interest. N. Bessonov and V. Volpert are external consultants
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix: A
1.1 A.1 Glossary
1.1.1 A.1.1 Abbreviations
- Met :
-
L-methionine
- MGL :
-
methionine-γ-lyase
- PN :
-
Pyridoxine
- PLP :
-
Pyridoxal phosphate
- CFSE :
-
Carboxyfluorescein diacetate succinimidyl ester
- Erymet :
-
Suspension of erythrocytes encapsulating MGL
- IG :
-
Intragastric
- IV :
-
Intravenous
- RBC :
-
Red Blood Cell
1.1.2 A.1.2 Symbols and Units
Concentration of methionine and PLP are expressed in μ M (μ mol.l− 1). Their doses are expressed in μ mol. The quantity of enzyme is expressed with its activity. The equations for MGL are not formulated in term of concentration because of the way used to measure the amount of enzyme. Amount of enzyme are expressed in units (Symbol U or UI). A unit of enzyme is the amount of enzyme that will catalyze the formation of 1 μ mol of product per minute under specifically defined conditions. The specific activity is the amount of enzyme activity per milligram of protein. For a given pure enzyme under a defined set of conditions, the specific activity is a constant [8]. For MGL, specific activity is 16.5 UI.mg− 1 unless otherwise specified in the text.
1.2 A.2 Signification of variables used in mathematical expression
1.3 A.3 Parameters for mice model
1.4 A.4 Fixed values for mice model
1.5 A.5 Identifiability of the models
We developed a biological model of the action of an enzyme (MGL) and its co-factor (PN) on the concentration of amino acid (MET) by comparison with preclinical data. We used Monolix, a software witch estimates the parameters of the model. This software is based on the maximum likelihood estimator of the population parameters, using the Stochastic Approximation Expectation Maximization (SAEM) algorithm.
We ensured the uniqueness of parameters recovered from observed data by carried out identifiability studies of our model.
Different methods have been proposed in the literature to test identifiability of nonlinear models. For [51], identifiability is the possibility to infer parameters from the data. The structural identifiability of compartmental models has been studied has been studied over a long period [50]. In [49], Groos and al. studied a more complex problem of identifiability of linear compartment models.
In our case, we consider the identifiability of each of the submodels.
Due to the complexity of the global model (see Fig. 2), we used a software to check the identifiability of the first three submodels. This software is DAISY (Differential Algebra for Identifiability of SYstems) [48]. The results given by the software are:
-
Submodels 1 and 2 are “locally identifiable”
-
Submodel 3 is “globally identifiable”.
We proof the identifiability of models 4 and 5 with the following calculus. Concerning the Emax model (submodels 4 and 5), with a simplify expression of equation we consider
For a given input and two sets of parameters \(\left (E^{1}_{max}, EC^{1}_{50}\right )\) and \(\left (E^{2}_{max}, EC^{2}_{50}\right )\), the output should be different. Indeed, if this is not the case, then we obtain Eq. A1 which cannot hold for any non-constant function Cinput(t). This contradiction proves the identifiability.
1.6 A.6 Modeling Met dietary intake (PE-TEDAC-MGL-012)
Protocol 12 provides plasma Met concentration with and without MGL administration, concentration of intraerythrocytic PLP and the activity of MGL at dates 15, 30, 45, 90, 180 and 1440 min for mice receiving an intravenous administration of MGL. PN is administered 5 days (administration date denoted: Day 0) after the injection of MGL. In what follows, time is expressed in minutes. The duration of protocol 12 is a single day (1440 min).
It was therefore possible to model the ordinary plasma level of methionine in mice (due to dietary intakes) with the data from the control population. Mice eat during night and stop their dietary intake during the day. We assume ten daily meals, even if mice have a continuous feeding.
Taking into account the composition of the food used for mice (SAFE A04), the daily intake of methionine varies between 8.4 and 16.8 mg per day per mouse. We assume a constant intake of 60 μ mol (10 mg per day per mouse) considering a molar mass of methionine equal to 149.21g.mol− 1. \(q_{MET_{1}}\), \(q_{MET_{2}}\) and \(q_{MET_{3}}\) (system of Eq. A2) are respectively the amount of methionine in the absorption, central and peripheral compartments. diet is the dietary intake of methionine.
1.6.1 Internal validation
1.7 A.7 Modeling of the intraerythrocytic PLP (PE-TEDAC-MGL-012)
Protocol PE-TEDAC-MGL-012 studied the pharmacokinetics of PLP (vitamin B6 oral uptake at day 0) following the MGL administration (at day -5). This protocol provides intraerythrocytic PLP concentrations at times 15, 30, 45, 90, 180 and 1440 min after an intragastric administration of PN for mice receiving PN, that is metabolized in PLP.

1.7.1 Internal validation
1.8 A.8 Modeling MGL kinetic (PE-TEDAC-MGL-013)
PLP is the co-factor of the enzyme (MGL) and is encapsulated with MGL in red blood cells to activate this enzyme. PLP progressively disappears, explaining an increasing inactive part of MGL.
Discursive scheme of MGL activity. Left: Total MGL. Right: Relationship between active and not active MGL. The first three in Eq. A4 describe this relationship
We first modeled the activity of MGL without administration of PN. We have denoted by “total activity”, the MGL activity measured with PLP against a range carried in biological matrix, “spontaneously active MGL” is the loaded in co-factor MGL and “non spontaneously active MGL” is the difference between the total and spontaneously active MGL. We used the system of Eq. A4 to describe this part of protocol 13 and the system (A2) to describe the methionine depletion after an IV of Erymet (first with PLP= 0).
1.8.1 Internal validation
1.8.2 A.8.1 Modeling pharmacodynamic of MGL
Choice of the model
At the time of its injection, the enzyme is in its active form (PLP is encapsulated with the enzyme). The global activity decreases rapidly and the remaining enzyme becomes more or less inactive depending on the amount of available PLP. We can observe the absence of hysteresis in experimental data and this could suggest a direct response model (see Fig. 16). Met as a function of encapsulated MGL in mice (experimental values from Erytech protocol, PE-TEDACMGL- 014). Mice receive PN two time per a day after Erymet administration. Consequently, a direct response model (Hill model) was chosen.
Modeling
The control population of protocol PE-TEDAC-MGL-013 provides activity of total MGL, activity of the activated MGL (by the initial encapsulation of PLP) and the part of the non activated MGL as a functions of time. The percentage of activated MGL decreases while the percentage of non activated MGL increases. Obviously, the activity of the total MGL decreases with time. We compared the activity of MGL with and without IG administrations of PN. We choose a direct model for the effect of PLP on MGL activity. To evaluate the importance of PN supplementation, we added Eq. A5 to the system (A4).
1.8.3 Internal validation
1.9 A.9 Modeling Met depletion (PE-TEDAC-MGL-012, 013)
The amount of the substrate (Met) decreases in the presence of the enzyme (MGL). Figure 18 shows a typical situation for which a direct response model is widely used. Consequently, we chose a direct response model for the effect of MGL on plasma Met.
After an administration of MGL and a supplementation with PN, the observed Met is described by
1.9.1 Internal validation
Development of the three submodels (modeling Eytech’s data from Protocols PE-TEDA-MGL-012, 13 and 14) can be seen in more details in Appendix A.7 for modeling of the intraerythrocytic PLP (PETEDAC-MGL-012), in Appendix A8 for enzime activity and in Appendix A.6 for modeling Met dietary intake (PE-TEDAC-MGL-012).
Appendix: B. E max model
The effect E is a measurable property whose nature depends on the tested drug. E may be the patient’s blood pressure drop or blood cholesterol levels, etc...
1.1 B.1 Action of the enzyme on the amino acid
In our case, the way to observe the effect of the drug is the measurement of blood methionine concentration and the effect E is its depletion. The differences between notation, due to the approaches differences, are summarized in the Table 5.
(a) Modeled plasma L-methionine concentration (control population in protocol 13: mice not receiving injection of MGL). (b) Modeled plasma L-methionine concentration in mice receiving injection of MGL in protocol 13. (c) Modeled plasma L-methionine concentration in mice receiving injection of MGL in protocol 12. (d) Modeled plasma L-methionine concentration in mice receiving injection of MGL in protocol 14. Y-axis: concentration of Met (μM), x-axis: time (min)
The classical Emax model of PK/PD modeling is established in the following calculus:
Let y the ratio of methionine receptors occupied by methionine-γ-lyase, i.e., the ratio of methionine which have reacted to form \([D\sim R]\).In equilibrium conditions, we may define [Rt], as the entire concentration of R As a consequence, \(y = \frac {[D\sim R]}{R_{t}} \text {, with } [R_{t}] = [R] + [D\sim R ]\)
We assume that the dissociation and association rates are equal when equilibrium state is reached,with dissociation rate = k2y and association rate = k1[D](1 − y), so k1[D](1 − y) = k2y
\(\text {On the other hand for an inhibition action, by definition: }y = \frac {E_{0} - E}{E_{max}}\)
with k1 = kinetic constant of association, k2 = kinetic constant of dissociation, kA = association constant = \(\frac {k_{1}}{k_{2}}\), kD = dissociation constant = \(\frac {k_{2}}{k_{1}}\).
Let kD = EC50. In what follows \(\frac {E_{max}}{E_{0}}\) is denoted Emax and is an unit-less quantity.
We obtained \(E = E_{0}\left (1 - E_{max} \frac {[D]}{[D] + EC_{50}}\right )\), i.e., \(C_{MET_{obs}} = C_{MET_{2}}\left (1-\frac {E_{max}^{MGL}{MGL_{obs}}}{EC_{50}^{MGL}+C_{MGL_{obs}}}\right )\) with the notation of the whole model.
We can notice without drug administration (\(C_{MGL_{a}} = 0\)), \(C_{MET_{obs}} = C_{MET_{2}}\).
1.2 B.2 Action of PN administration on encapsulated methionine-γ-lyase
PN becomes PLP, witch is the co-factor of the enzyme consequently the effect of the drug (PLP) is the increase in activity of encapsulated methionine-γ-lyase, then \(E = C_{MGL_{a}}^{PLP}\) and the baseline effect prior to drug administration \(E_{0} = C_{MGL_{a}}\), i.e., E0 is the activity without PLP administration.
MGLa and MGLi are active and inactive encapsulated methionine-γ-lyase amount in the central compartment (the administered red cells loaded with the enzyme). MGLp is the amount of enzyme in the peripheral compartment.
In order to simplify the reading of Eq. B3, \(C_{MGL_{a}}\) is denoted [G] and CPLP is denoted [P].
equilibrium equation, \(\frac {[P\sim G ]}{dt} =0 \text {, } k_{1} [P][G] = k_{2} [P\sim G ]\)Let y, the number of agonist occupied receptors.We assume that the dissociation and association rates are equal, with dissociationrate = k2y,association rate = k1[P](1 − y),so k1[P](1 − y) = k2y
We obtained \(E = E_{0}\left (1 + E_{max} \frac {[P]}{[P] + EC_{50}}\right )\) i.e., \(C_{MGL_{a}}^{PLP} = C_{MGL_{a}}\left (1+\frac {E_{{max}^{PLP}}^{a} C_{PLP}}{EC_{50}^{PLP}+C_{PLP}}\right )\) with the notation of the whole model.
Appendix: C Computational aspects and parameters assessment
The global model PK/PD has been implemented with MLXTRAN code (the language for describing models and treatment design of the Monolix software, a software for nonlinear mixed effects modeling) to assess parameters, their values are given in Table 1 and some parameters are assessed by nonlinear multiple regression method implemented in Scilab. The output variables were total MGL, active MGL, non active MGL, PLP and Met. For each variable, we used a constant error. The standard errors were obtained by linearization, with conditional modes for individual parameters and were obtained by linearization for Log-likelihood.
Simulations of PK/PD model have been made by writing code with Scilab, the system of ordinary differential equations has been solved by the ordinary differential equation solver “ode”, specific code has been written to solve the system taken into account various frequency of administrations.
The tumor model is implemented in C++ with the operating system Ubuntu and a graphical library wxWidget [39] included in specific software developed [40, 41]. Cell divisions are model as follows: during cell cycle, cell radius grows linearly. It becomes twice the initial radius at the end of the cycle. Then, each mother cell is replaced by two daughter cells. Geometrically, two circles with the initial radius are located inside the circle with a twice larger radius. The direction of cell division, that is the direction of the line connecting the centers of daughter cells is random [39]. On a computer view, a cell is a set of parameters. In order not to have a predetermine cell behavior, these values can vary with time and cells location. Each cell has its own initial and critical values and coefficients of equations considered in the model. Their fate depend on the computation results done during their whole life. To increase legibility of this representation, cells can not overlap. It is therefore necessary to consider their positions and displacements. A more complete approach of cells motion in hybrid models can be found in [39, 41,42,43,44].
The equations of the model are solved with an algorithm involving the spatial discretization of a computational domain and discretization of the time of simulation, i.e., each iteration of the computation corresponds to a time step Δt. In numerical simulations, Eq. 6 is solved by a finite difference method with Thomas algorithm and an alternative direction method. Neumann (no-flux) boundary conditions are considered as the boundary of the rectangular domain (L x L) [39, 44]. Constant space and time steps are used. Equation 7 is solved by Euler method.
In Eq. 6, the consumption rate of methionine by cancer cells is calculated as follows (see [45]): A local cell concentration c is computed as total cell area inside each square of the grid. For simplicity of calculation of cell concentration we make an assumption that cell is square (see Fig. 20). If one square grid of numerical scheme is completely inside of the cell, then concentration is equal
Otherwise, cell concentration is
Next, we distribute the concentration between the nodes dividing c1 and c2 by 4. Then we compute the values of the concentration at the nodes of the numerical grid corresponding to Eq. 6.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eymard, N., Bessonov, N., Volpert, V. et al. Pharmacokinetic/pharmacodynamic model of a methionine starvation based anti-cancer drug. Med Biol Eng Comput 61, 1697–1722 (2023). https://doi.org/10.1007/s11517-023-02786-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02786-2