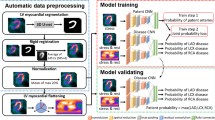
Abstract
Myocardial ischemia diagnosis with CT perfusion imaging (CTP) is important in coronary artery disease management. Traditional analysis procedure is time-consuming and error-prone due to the semi-manual and operator-dependent nature. To improve the diagnostic performance, a deep learning-based, fully automatic, and clinical-ready framework was developed. Two collaborating deep learning networks including a 3D U-Net for left ventricle segmentation and a CNN for anatomical landmarks detection were trained on 276 subjects. With our processing framework, the 17-segment left ventricular model was automatically generated conformed to the clinical standard. Myocardial blood flow computed by commercial software was extracted within each segment and visualized against the bull’s eye plot. The performance was validated on another 45 subjects. Coronary angiography and invasive fractional flow reserve measurements were also performed in these patients to serve as the gold standard for myocardial ischemia diagnosis. As a result, the diagnostic accuracy for our method was 81.08%, much higher than that for commercially available CTP analysis software (56.75%), and our method demonstrated a higher consistency (Kappa coefficient 0.759 vs. 0.585). Besides, the average processing time of our method was much lower (30 ± 10.5 s/subject vs. over 30 min/subject). In conclusion, the proposed deep learning-based framework could be a promising tool for assisting CTP analysis.
Graphical Abstract








Similar content being viewed by others
References
Yamamuro M, Tadamura E, Kubo S et al (2005) Cardiac functional analysis with multi–detector row CT and segmental reconstruction algorithm: comparison with echocardiography, SPECT, and MR imaging. Radiol 234:381–390. https://doi.org/10.1148/radiol.2342031271
American Heart Association Writing Group on Myocardial Segmentation and Registration for Cardiac Imaging, Cerqueira MD, Weissman NJ et al (2002) Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circ 105:539–542. https://doi.org/10.1161/hc0402.102975
Ebersberger U, Marcus RP, Schoepf UJ et al (2014) Dynamic CT myocardial perfusion imaging: performance of 3D semi-automated evaluation software. Eur Radiol 24:191–199. https://doi.org/10.1007/s00330-013-2997-5
Afshin M, Ben Ayed I, Punithakumar K et al (2014) Regional assessment of cardiac left ventricular myocardial function via MRI statistical features. IEEE Trans Med Imaging 33:481–494. https://doi.org/10.1109/TMI.2013.2287793
Liang Xi, Garnavi R, Wail S et al (2015) Automatic segmentation of the left ventricle into 17 anatomical regions in cardiac MR imaging. 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, Milan, pp 6531–6535
Bai W, Peressutti D, Parisot S et al (2016) Beyond the AHA 17-segment model: motion-driven parcellation of the left ventricle. In: Camara O, Mansi T, Pop M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, Imaging and Modelling Challenges, pp 13–20
Romaguera LV, Romero FP, Fernandes Costa Filho CF, Fernandes Costa MG (2018) Myocardial segmentation in cardiac magnetic resonance images using fully convolutional neural networks. Biomed Signal Process Control 44:48–57. https://doi.org/10.1016/j.bspc.2018.04.008
Curiale AH, Colavecchia FD, Kaluza P (2017) Automatic myocardial segmentation by using a deep learning network in cardiac MRI. In: 2017 XLIII Latin American Computer Conference (CLEI). IEEE, Cordoba, pp 1–6. https://doi.org/10.1109/CLEI.2017.8226420
Kuang M, Wu Y, Alonso-Álvarez D (2021) Three-dimensional embedded attentive RNN (3D-EAR) segmentor for left ventricle delineation from myocardial velocity mapping. https://doi.org/10.48550/arXiv.2104.13214
Isensee F, Jaeger PF, Kohl SAA et al (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18:203–211. https://doi.org/10.1038/s41592-020-01008-z
Çiçek Ö, Abdulkadir A, Lienkamp SS (2016) Medical Image Computing and Computer-Assisted Intervention– MICCAI 2016, Lecture Notes in Computer Science. In: Ourselin S, Joskowicz L, Sabuncu MR MR, Unal G, Wells W (eds) D U-Net: learning dense volumetric segmentation from sparse annotation, in: Springer International Publishing, Cham, pp 424–432.https://doi.org/10.1007/978-3-319-46723-8_49
Wang L, Wang C, Sun Z, Chen S (2020) An improved dice loss for pneumothorax segmentation by mining the information of negative areas. IEEE Access 8:167939–167949. https://doi.org/10.1109/ACCESS.2020.3020475
Kingma DP, Ba J (2017) Adam: a method for stochastic optimization. https://doi.org/10.48550/arXiv.1412.6980
Schiller NB, Shah PM, Crawford M et al (1989) Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. J Am Soc Echocardiogr 2:358–367. https://doi.org/10.1016/S0894-7317(89)80014-8
Reza A, Sengupta AS (2017) Least square ellipsoid fitting using iterative orthogonal transformations. Appl Math Comput 314:349–359.https://doi.org/10.1016/j.amc.2017.07.025
Milletari F, Navab N, Ahmadi SA (2016) V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 016 Fourth International Conference on 3D Vision (3DV). IEEE, Stanford, CA, USA, pp 565–571. https://doi.org/10.1109/3DV.2016.79
Liu D, Hu K, Nordbeck P et al (2016) Longitudinal strain bull’s eye plot patterns in patients with cardiomyopathy and concentric left ventricular hypertrophy. Eur J Med Res 21:21. https://doi.org/10.1186/s40001-016-0216-y
Adiputra Y, Chen S-L (2015) Clinical relevance of coronary fractional flow reserve: art-of-state. Chin Med J 128:1399–1406.https://doi.org/10.4103/0366-6999.156805
Coenen A, Rossi A, Lubbers MM et al (2017) Integrating CT myocardial perfusion and CT-FFR in the work-up of coronary artery disease. JACC: Cardiovasc Imaging 10:760–770. https://doi.org/10.1016/j.jcmg.2016.09.028
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015
Grinias E, Tziritas G (2018) Fast fully-automatic cardiac segmentation in MRI using MRF model optimization, substructures tracking and B-spline smoothing. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 91–100
Jang Y, Hong Y, Ha S et al (2018) Automatic segmentation of LV and RV in cardiac MRI. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 161–169
Khened M, Alex V, Krishnamurthi G (2018) Densely connected fully convolutional network for short-axis cardiac cine MR image segmentation and heart diagnosis using random forest. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 140–151
Patravali J, Jain S, Chilamkurthy S (2018) 2D–3D fully convolutional neural networks for cardiac MR segmentation. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 130–139
Yang X, Bian C, Yu L et al (2018) Class-balanced deep neural network for automatic ventricular structure segmentation. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 152–160
Isensee F, Jaeger P, Full PM et al (2018). Automatic cardiac disease assessment on cine-MRI via time-series segmentation and domain specific features. https://doi.org/10.1007/978-3-319-75541-0
Yang X, Bian C, Yu L et al (2018) 3D Convolutional networks for fully automatic fine-grained whole heart partition. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 181–189
Payer C, Štern D, Bischof H, Urschler M (2018) Multi-label whole heart segmentation using CNNs and anatomical label configurations. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical Atlases and Computational Models of the Heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 190–198
Yang X, Bian C, Yu L et al (2018) Hybrid loss guided convolutional networks for whole heart parsing. In: Pop M, Sermesant M, Jodoin P-M et al (eds) Statistical atlases and computational models of the heart. Springer International Publishing, Cham, ACDC and MMWHS Challenges, pp 215–223
Caiani E, Toledo E, MacEneaney P et al (2006) Automated interpretation of regional left ventricular wall motion from cardiac magnetic resonance images. J Cardiovasc Magn Reson 8:427–433. https://doi.org/10.1080/10976640600599486
Lekadir K, Keenan NG, Pennell DJ, Yang G (2011) An inter-landmark approach to 4-D shape extraction and interpretation: application to myocardial motion assessment in MRI. IEEE Trans Med Imaging 30:52–68. https://doi.org/10.1109/TMI.2010.2060490
Lu Y, Radau P, Connelly K et al (2009) Pattern recognition of abnormal left ventricle wall motion in cardiac MR. In: Yang G-Z, Hawkes D, Rueckert D et al (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2009. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 750–758
Punithakumar K, Ben Ayed I, Islam A et al (2013) Regional heart motion abnormality detection: an information theoretic approach. Med Image Anal 17:311–324. https://doi.org/10.1016/j.media.2012.11.007
Funding
This study was supported by the Shenzhen Science and Technology Program (grant number KQTD2016112809330877), the Clinical Research Center of Shandong University (grant number 2020SDUCRCB005), the Key Research and Development Program of Shandong Province (grant number 2020ZLYS05), and the Shenyang Science and Technology Plan Project (grant number 20–205-4–014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Dr. Schoepf receives institutional research support and/or personal fees for speaking and consulting from Bayer, Bracco, GE, Guerbet, HeartFlow Inc., Keya Medical, and Siemens Healthineers. MA, JL, XX, RHS, KC, QS, YL, PZ, and ZL have no potential competing interests to declare.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
An, M., Li, J., Xu, X. et al. A deep learning-based fully automatic and clinical-ready framework for regional myocardial segmentation and myocardial ischemia evaluation. Med Biol Eng Comput 61, 1507–1520 (2023). https://doi.org/10.1007/s11517-023-02798-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02798-y