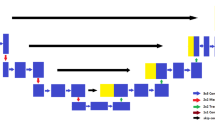
Abstract
In hospitals and pathology, observing the features and locations of brain tumors in Magnetic Resonance Images (MRI) is a crucial task for assisting medical professionals in both treatment and diagnosis. The multi-class information about the brain tumor is often obtained from the patient’s MRI dataset. However, this information may vary in different shapes and sizes for various brain tumors, making it difficult to detect their locations in the brain. To resolve these issues, a novel customized Deep Convolution Neural Network (DCNN) based Residual-Unet (ResUnet) model with Transfer Learning (TL) is proposed for predicting the locations of the brain tumor in an MRI dataset. The DCNN model has been used to extract the features from input images and select the Region Of Interest (ROI) by using the TL technique for training it faster. Furthermore, the min-max normalizing approach is used to enhance the color intensity value for particular ROI boundary edges in the brain tumor images. Specifically, the boundary edges of the brain tumors have been detected by utilizing Gateaux Derivatives (GD) method to identify the multi-class brain tumors precisely. The proposed scheme has been validated on two datasets namely the brain tumor, and Figshare MRI datasets for detecting multi-class Brain Tumor Segmentation (BTS).The experimental results have been analyzed by evaluation metrics namely, accuracy (99.78, and 99.03), Jaccard Coefficient (93.04, and 94.95), Dice Factor Coefficient (DFC) (92.37, and 91.94), Mean Absolute Error (MAE) (0.0019, and 0.0013), and Mean Squared Error (MSE) (0.0085, and 0.0012) for proper validation. The proposed system outperforms the state-of-the-art segmentation models on the MRI brain tumor dataset.
Graphical abstract















Similar content being viewed by others
Data Availability
The data supporting the findings of this manuscript are available at this link: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 for Figshare dataset and link: https://www.kaggle.com/datasets/nikhilroxtomar/ brain-tumor-segmentation for braintumor dataset.
References
Tran ST, Cheng CH, Nguyen TT, Le MH, Liu DG (2021) TMD-Unet: Triple-Unet with Multi-Scale Input Features and Dense Skip Connection for Medical Image Segmentation. Healthcare 9(1):54–73. https://doi.org/10.3390/healthcare9010054
NH Narayana Health, Health for all, All for health, NH CARES Brain Tumour, Types, Risk Factors, Symptoms, and Surgery. Available online: https://www.narayanahealth.org/brain-tumour
Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal 43:98–111. https://doi.org/10.1016/j.media.2017.10.002
Bauer S, Wiest R, Nolte LP, Reyes M (2013) A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol 58(13):97–140
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78. https://doi.org/10.1016/j.media.2016.10.004
Subashini MM, Sahoo SK, Sunil V, Easwaran S (2016) A non-invasive methodology for the grade identification of astrocytoma using image processing and artificial intelligence techniques. Expert Syst Appl 43:186–196. https://doi.org/10.1016/j.eswa.2015.08.036
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251. https://doi.org/10.1109/TMI.2016.2538465
Iqbal S, Ghani MU, Saba T, Rehman A (2018) Brain tumor segmentation in multi-spectral MRI using convolutional neural networks (CNN). Microsc Res Tech 81(4):419–427. https://doi.org/10.1002/jemt.22994
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Van Leemput K (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Gooya A, Pohl KM, Bilello M, Cirillo L, Biros G, Melhem ER, Davatzikos C (2012) GLISTR: glioma image segmentation and registration. IEEE Trans Med Imaging 31(10):1941–1954. https://doi.org/10.1109/TMI.2012.2210558
Nadipineni H (2020) Method to classify skin lesions using dermoscopic images. arXiv preprint arXiv:2008.09418. 153:113419. https://doi.org/10.1016/j.eswa.2020.113419
Lee B, Yamanakkanavar N, Choi JY (2020) Automatic segmentation of brain MRI using a novel patch-wise U-net deep architecture. PLoS ONE 15(8):e0236493. https://doi.org/10.1371/journal.pone.0236493
Song Y, Ren S, Lu Y, Fu X, Wong KK (2022) Deep learning-based automatic segmentation of images in cardiac radiography: A promising challenge. Comput Methods Prog Biomed 220:106821. https://doi.org/10.1016/j.cmpb.2022.106821
Homayounieh F, Singh R, Nitiwarangkul C, Lades F, Schmidt B, Sedlmair M, Kalra MK (2020) Semiautomatic segmentation and radiomics for dual-energy CT: a pilot study to differentiate benign and malignant hepatic lesions. Am J Roentgenol 215(2):398–405. https://doi.org/10.2214/AJR.19.22164
Lima PV, de MS Veras R, Vogado LH, Portela HM, de Almeida JD, Aires KR, Leite D, (2020) A semiautomatic segmentation approach to corneal lesions. Comput Electr Eng 84:106625. https://doi.org/10.1016/j.compeleceng.2020.106625
Zhou Y, Huang W, Dong P, Xia Y, Wang S (2019) D-UNet: a dimension-fusion U shape network for chronic stroke lesion segmentation. IEEE/ACM Trans Comput Biol Bioinforma 18(3):940–950. https://doi.org/10.1109/TCBB.2019.2939522
Wu G, Chen Y, Wang Y, Yu J, Lv X, Ju X, Chen Z (2017) Sparse representation-based radiomics for the diagnosis of brain tumors. IEEE Trans Med Imaging 37(4):893–905. https://doi.org/10.1109/TMI.2017.2776967
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal 39:640–651
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234-241. https://doi.org/10.1007/978-3-319-24574-4_28
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2017) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans Pattern Anal Mach Intell 40(4):834–848. https://doi.org/10.1109/TPAMI.2017.2699184
Badrinarayanan V, Kendall A, Cipolla R (2017) SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495. https://doi.org/10.1109/TPAMI.2016.2644615
Murmu A, Kumar P (2021) Deep learning model-based segmen-tation of medical diseases from MRI and CT images. In: TENCON 2021 IEEE Region 10 Conference (TENCON), pp 608-613. https://doi.org/10.1109/TENCON54134.2021.9707278
Abdollahi A, Pradhan B, Alamri A (2020) VNet: An end-to-end fully convolutional neural network for road extraction from high-resolution remote sensing data. IEEE Access 8:179424–179436. https://doi.org/10.1109/ACCESS.2020.3026658
Ding Y, Zheng W, Geng J, Qin Z, Choo KKR, Qin Z, Hou X (2021) MVFusFra: A Multi-View Dynamic Fusion Framework for Multimodal Brain Tumor Segmentation. IEEE J Biomed Health Inform 26(4):1570–1581. https://doi.org/10.1109/JBHI.2021.3122328
Yu B, Zhou L, Wang L, Yang W, Yang M, Bourgeat P, Fripp J (2021) SA-LuT-Nets: learning sample-adaptive intensity lookup tables for brain tumor segmentation. IEEE Trans Med Imaging 40(5):1417–1427. https://doi.org/10.1109/TMI.2021.3056678
Shaikh M, Anand G, Acharya G, Amrutkar A, Alex V, Krishnamurthi G (2017) Brain tumor segmentation using dense fully convolutional neural network. In: international MICCAI brainlesion workshop. Springer, pp 309-319
Kleesiek J, Urban G, Hubert A, Schwarz D, Maier-Hein K, Bendszus M, Biller A (2016) Deep MRI brain extraction: A 3D convolutional neural network for skull stripping. NeuroImage 129:460–469. https://doi.org/10.1016/j.neuroimage.2016.01.024
Moeskops P, Viergever MA, Mendrik AM, De Vries LS, Benders MJ, Išgum I (2016) Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging 35(5):1252–1261. https://doi.org/10.1109/TMI.2016.2548501
Attallah O, Sharkas MA, Gadelkarim H (2020) Deep learning techniques for automatic detection of embryonic neurodevelopmental disorders. Diagnostics 10(1):27–49. https://doi.org/10.3390/diagnostics10010027
Stadlbauer A, Marhold F, Oberndorfer S, Heinz G, Buchfelder M, Kinfe TM, Meyer-Bäse A (2022) Radiophysiomics: Brain Tumors Classification by Machine Learning and Physiological MRI Data. Cancers 14(10):2363–2385. https://doi.org/10.3390/cancers14102363
Attallah O, Sharkas MA, Gadelkarim H (2019) Fetal brain abnormality classification from MRI images of different gestational age. Brain Sci 9(9):231–252. https://doi.org/10.3390/brainsci9090231
Aamir M, Rahman Z, Dayo ZA, Abro WA, Uddin MI, Khan I, Hu Z (2022) A deep learning approach for brain tumor classification using MRI images. Comput Electr Eng 101:108105. https://doi.org/10.1016/j.compeleceng.2022.108105
Jun C (2017) Brain tumor dataset. Available: https://figshare.com/articles/brain_tumor_dataset/1512427. Accessed 16 Aug 2022
Tomar N (2022) Brain Tumor Segmentation dataset. Available: https://www.kaggle.com/datasets/nikhilroxtomar/brain-tumor-segmentation. Accessed 10 Dec 2022
Kumar P, Agrawal A (2013) CUDA-based interactive volume rendering of 3D medical data. Springer international Conference on intelligent interactive technologies and multimedia. Springer, pp 123-132. https://doi.org/10.1007/978-3-642-37463-0_11
Kibriya H, Masood M, Nawaz M, Nazir T (2022) Multiclass classification of brain tumors using a novel CNN architecture. Multimed Tools Appl 81:29847–29863. https://doi.org/10.1007/s11042-022-12977-y
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251. https://doi.org/10.1109/TMI.2016.2538465
Anaraki AK, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 39(1):63–74. https://doi.org/10.1016/j.bbe.2018.10.004
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251. https://doi.org/10.1109/TMI.2016.2538465
Kass M, Witkin A, Terzopoulos D (1988) Snakes: Active contour models. Int J Comput Vis 1(4):321–331. https://doi.org/10.1007/BF00133570
Hendrycks D, Gimpel K (2016) Gaussian error linear units (ge-lus). arXiv preprint arXiv:1606.08415. https://doi.org/10.48550/arXiv.1606.08415
Moeskops P, Viergever MA, Mendrik AM, De Vries LS, Benders MJ, Išgum I (2016) Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging 35(5):1252–1261. https://doi.org/10.1109/TMI.2016.2548501
Oktay O, Schlemper J, Folgoc L L, Lee M, Heinrich M, Misawa K, Rueckert D (2018). Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999. https://doi.org/10.48550/arXiv.1804.03999
Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y, Wu J (2020) Unet 3+: A full-scale connected unet for medical image segmentation. In: ICASSP 2020 IEEE international conference on acoustics, speech and signal processing (ICASSP), pp 1055-1059. https://doi.org/10.1109/ICASSP40776.2020.9053405
Diakogiannis FI, Waldner F, Caccetta P, Wu C (2020) ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J Photogramm Remote Sens 162:94–114. https://doi.org/10.1016/j.isprsjprs.2020.01.013
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Zhou Y (2021) Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306. https://doi.org/10.48550/arXiv.2102.04306
Lou A, Guan S, Loew M (2021) DC-UNet: rethinking the U-Net architecture with dual channel efficient CNN for medical image segmentation. Med Imaging 11596:758–768. https://doi.org/10.1117/12.2582338
Zhang Z, Duan C, Lin T, Zhou S, Wang Y, Gao X (2020) GVFOM: a novel external force for active contour based image segmentation. Inf Sci 506:1–18. https://doi.org/10.1016/j.ins.2019.08.003
Ayub M, Ghazanfar MA, Khan T, Saleem A (2020) An effective model for Jaccard coefficient to increase the performance of collaborative filtering. Arab J Sci Eng 45(12):9997–10017. https://doi.org/10.1007/s13369-020-04568-6
Kofler F, Ezhov I, Isensee F, Balsiger F, Berger C, Koerner M, Menze B H (2021) Are we using appropriate segmentation metrics? Identifying correlates of human expert perception for CNN training beyond rolling the DICE coefficient. arXiv preprint arXiv:2103.06205
Ravuri S, Vinyals O (2019) Classification accuracy score for conditional generative models. Adv Neural Inf Process Syst 32
Chicco D, Jurman G (2020) The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21(1):1–13. https://doi.org/10.1186/s12864-019-6413-7
Ruby U, Yendapalli V (2020) Binary cross entropy with deep learning technique for image classification. Int J Adv Trends Comput Sci Eng 9(10):5393-5397. https://doi.org/10.30534/ijatcse/2020/175942020
Karunasingha DSK (2022) Root mean square error or mean absolute error? Use their ratio as well. Inf Sci 585:609–629. https://doi.org/10.1016/j.ins.2021.11.036
Hodson TO, Over TM, Foks SS (2021) Mean squared error, deconstructed. J Adv Model Earth Syst 13(12):e2021MS002681. https://doi.org/10.1029/2021MS002681
Bottou L, Bengio Y, Le Cun Y (1997) Global training of document processing systems using graph transformer networks. In: proceedings of IEEE computer society conference on computer vision and pattern recognition, pp 489-494. https://doi.org/10.1109/CVPR.1997.609370
Lafferty J D, McCallum A, Pereira F C N (2001) Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In: Proceeding 18th international conference in machine learning, USA, pp 282-289
Tong X, Xu X, Huang SL, Zheng L (2021) A Mathematical Framework for Quantifying Transferability in Multi-source Transfer Learning. Adv Neural Inf Process Syst 34:26103-26116
Hasan AM, Meziane F, Aspin R, Jalab HA (2016) Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry 8(11):132–152. https://doi.org/10.3390/sym8110132
Abdel-Maksoud E, Elmogy M, Al-Awadi R (2015) Brain tumor segmentation based on a hybrid clustering technique. Egypt Inform J 16(1):71–81. https://doi.org/10.1016/j.eij.2015.01.003
Chandra SK, Bajpai MK (2020) Brain tumor detection and segmentation using mesh-free super-diffusive model. Multimed Tools Appl 79(3):2653–2670. https://doi.org/10.1007/s11042-019-08374-7
Zhou T, Canu S, Vera P, Ruan S (2021) Latent correlation representation learning for brain tumor segmentation with missing MRI modalities. IEEE Trans Image Process 30:4263–4274. https://doi.org/10.1109/TIP.2021.3070752
Ma J, Wang D, Wang XP, Yang X (2021) A characteristic function-based algorithm for geodesic active contours. SIAM J Imaging Sci 14(3):1184–1205. https://doi.org/10.1137/20M1382817
Funding
No funding provided from any source is used for this research in this manuscript.
Author information
Authors and Affiliations
Contributions
Anita Murmu (AM) designed the Environmental setup platform, conducted the experiments, and performed statistical analyses. Where Piyush Kumar (PiK) wrote the abstract section and literature survey. AM and PiK wrote the first draft of the manuscript. AM and PiK contributed to the investigation and framing of the results. PiK edited the first draft of this paper. Both authors participated in reviewing and approved the final version of the manuscript. Anita Murmu and Piyush Kumar, both authors contributed equally to this work.
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors. Therefore, this section is not applicable to this paper.
Conflict of interest/Conflict of interest:
Both authors declare that he or she has no conflict of interest with respect to the research, authorship, and/or publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendix A: Snakes
The geodesic active counter (GAC) model is used to retrieve objects from images (also called Snakes). GAC models are given in Eq. (30) by author [63]. Let, \(W_{k}=W\) is function of counter interacts of the image, where k=1,2,...,k.
Equation (29) has been describe in below Eq. (30)
where, \(\Gamma ^{t} = \left\{ x \epsilon D:\Phi (q,t)=0 \right\} \). The idea is that g is small near object boundaries. The first term describes the external energy which pushes the contours toward the object’s border, whilst the second and third terms are associated with the internal energy that regulates the smoothing of the contours. So, getting Eq. (2).
where \(g(\bigtriangledown _{q}) = \frac{1}{1 + \gamma \mid \bigtriangledown _{q} \mid }\)
As a result, the image and the prospective function \(W:D \rightarrow R\) interact since it is a lower limit for some threshold and I(x) is the \(\varepsilon (\Gamma )\).
Therefore, getting Eq. (3), \(W(x)=\frac{1}{1+ \left\| \bigtriangledown Z \sigma *I(x))\right\| ^{2}}\)
Now, the result reduces \(\varepsilon \) value which attracts \(\Gamma \) to the margins. The descent flow is a fourth order nonlinear parabolic equation (4) we get.
Appendix B: Brain tumor boundary detection
The simplification of Eq (8). Assume that \(C (z,t): D \times [0, \alpha ) \rightarrow \mathbb {R}^{n}\) is a collection of planar arcs in the given image, and that \(\sigma _{1}, \sigma _{2}\) are the mean intensities of I both outside and inside C(z,t) respectively. Then
where, \(\omega ^{c}\) is exterior, and \(\omega \) is interior of C(z,t). Based on the distance, C is able to distinguish between the background and foreground areas.
Sup is Sup is C on \(E^{1,2}(D)= \left\{ C \epsilon L^{2}(D);\bigtriangledown C \epsilon L^{2}(D)\right\} \) We rephrase Eq. (33) as a minimization issue in order to increase the applicability of its skills to a wide range of images. We also add an edge function f to enhance its capability for detection of boundary.
We include a regularization to enhance Eq. (34) even further \(L = \oint _{c} dt\) to get Eq. (8) \(E(z, \omega ) = -\frac{1}{2} f(z) (\sigma _{1} - \sigma _{2})^{2} + \beta \oint _\partial \omega dt \)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Murmu, A., Kumar, P. A novel Gateaux derivatives with efficient DCNN-Resunet method for segmenting multi-class brain tumor. Med Biol Eng Comput 61, 2115–2138 (2023). https://doi.org/10.1007/s11517-023-02824-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02824-z