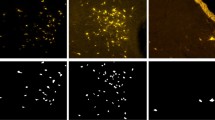
Abstract
Medical image processing has become increasingly important in recent years, particularly in the field of microscopic cell imaging. However, accurately counting the number of cells in an image can be a challenging task due to the significant variations in cell size and shape. To tackle this problem, many existing methods rely on deep learning techniques, such as convolutional neural networks (CNNs), to count cells in an image or use regression counting methods to learn the similarities between an input image and a predicted cell image density map. In this paper, we propose a novel approach to monitor the cell counting process by optimizing the loss function using the optimal transport method, a rigorous measure to calculate the difference between the predicted count map and the dot annotation map generated by the CNN. We evaluated our algorithm on three publicly available cell count benchmarks: the synthetic fluorescence microscopy (VGG) dataset, the modified bone marrow (MBM) dataset, and the human subcutaneous adipose tissue (ADI) dataset. Our method outperforms other state-of-the-art methods, achieving a mean absolute error (MAE) of 2.3, 4.8, and 13.1 on the VGG, MBM, and ADI datasets, respectively, with smaller standard deviations. By using the optimal transport method, our approach provides a more accurate and reliable cell counting method for medical image processing.
Graphical abstract






Similar content being viewed by others
References
Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, Böhm A, Deubner J, Jäckel Z, Seiwald K et al (2019) U-net: deep learning for cell counting, detection, and morphometry. Nat Methods 16(1):67–70
Zhang C, Li H, Wang X, Yang X (2015) Cross-scene crowd counting via deep convolutional neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp 833–841
Laradji IH, Rostamzadeh N, Pinheiro PO, Vazquez D, Schmidt M (2018) Where are the blobs: counting by localization with point supervision. In Springer, Cham
Polley M-YC, Leung SC, McShane LM, Gao D, Hugh JC, Mastropasqua MG, Viale G, Zabaglo LA, Penault-Llorca F, Bartlett JM et al (2013) An international Ki67 reproducibility study. J Natl Cancer Inst 105(24):1897–1906
Xie W, Noble JA, Zisserman A (2018) Microscopy cell counting and detection with fully convolutional regression networks. Comput Methods Biomech Biomed Eng: Imaging & Visualization 6(3):283–292
He S, Minn KT, Solnica-Krezel L, Anastasio M, Li H (2019) Automatic microscopic cell counting by use of deeply-supervised density regression model. In Medical Imaging 2019: Digital Pathology, vol 10956, pp 121–128 SPIE
Guo Y, Stein J, Wu G, Krishnamurthy A (2019) SAU-Net: a universal deep network for cell counting. In Proceedings of the 10th ACM international conference on bioinformatics, computational biology and health informatics, pp 299–306
Wang Z, Yin Z (2021) Cell counting by a location-aware network. In International workshop on machine learning in medical imaging, Springer pp 120–129
Jiang N, Yu F (2020) A cell counting framework based on random forest and density map. Appl Sci 10(23):8346
Barinova O, Lempitsky V, Kholi P (2012) On detection of multiple object instances using Hough transforms. IEEE Trans Pattern Anal Mach Intell 34(9):1773–1784
Arteta C, Lempitsky V, Noble JA, Zisserman A (2012) Learning to detect cells using non-overlapping extremal regions. In International conference on medical image computing and computer-assisted intervention, Springer pp 348–356
Xing F, Su H, Neltner J, Yang L (2013) Automatic Ki-67 counting using robust cell detection and online dictionary learning. IEEE Transact Biomed Eng 61(3):859–870
Arteta C, Lempitsky V, Noble JA, Zisserman A (2016) Detecting overlapping instances in microscopy images using extremal region trees. Med Image Anal 27:3–16
Lempitsky V, Zisserman A (2010) Learning to count objects in images. Adv Neural Inf Process Syst 23
Xie Y, Xing F, Kong X, Su H, Yang L (2015) Beyond classification: structured regression for robust cell detection using convolutional neural network. In International conference on medical image computing and computer-assisted intervention, Springer pp 358–365
Paul Cohen J, Boucher G, Glastonbury CA, Lo HZ, Bengio Y (2017) Count-ception: counting by fully convolutional redundant counting. In Proceedings of the IEEE international conference on computer vision workshops, pp 18–26
Xu M, Hu M, Zhang Y, Zhou Y (2021) DAU-Net: a regression cell counting method. In ISCTT 2021; 6th International conference on information science, computer technology and transportation, VDE pp 1–6
Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J (2013) Mitosis detection in breast cancer histology images with deep neural networks. In International conference on medical image computing and computer-assisted intervention, Springer pp 411–418
Fiaschi L, Köthe U, Nair R, Hamprecht FA (2012) Learning to count with regression forest and structured labels. In Proceedings of the 21st international conference on pattern recognition (ICPR2012), IEEE pp 2685–2688
Arteta C, Lempitsky V, Noble JA, Zisserman A (2014) Interactive object counting. In European conference on computer vision, Springer pp 504–518
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Seguí S, Pujol O, Vitria J (2015) Learning to count with deep object features. In Proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp 90–96
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In International conference on medical image computing and computer-assisted intervention, Springer pp 234–241
Wan J, Chan A (2019) Adaptive density map generation for crowd counting. In Proceedings of the IEEE/CVF international conference on computer vision, pp 1130–1139
Ma Z, Wei X, Hong X, Gong Y (2019) Bayesian loss for crowd count estimation with point supervision. In Proceedings of the IEEE/CVF international conference on computer vision, pp 6142–6151
Wang B, Liu H, Samaras D, Nguyen MH (2020) Distribution matching for crowd counting. Adv Neural Inf Process Syst 33:1595–1607
Wan J, Liu Z, Chan AB (2021) A generalized loss function for crowd counting and localization. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 1974–1983
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Villani C (2009) Optimal transport: old and new, vol 338. Springer, Berlin
Rubner Y, Tomasi C, Guibas LJ (2000) The earth mover’s distance as a metric for image retrieval. Int J Comput Vision 40(2):99–21
Chen P, Gui C (2013) Alpha divergences based mass transport models for image matching problems. Inverse Problems & Imaging 5(3):551–590
Gibbs AL, Su FE (2010) On choosing and bounding probability metrics. Int Stat Rev 70(3):419–435
Kantorovich LV (2006) On the translocation of masses. J Math Sci 133(4):1381–1382
Bregman LM (1967) Proof of the convergence of Sheleikhovskii’s method for a problem with transportation constraints. USSR Comput Math Math Phys 7(1):191–204
Sinkhorn R (1974) Diagonal equivalence to matrices with prescribed row and column sums. ii. Proc Am Math Soc 45(2):195–198
Kalantari B, Khachiyan L (1993) On the rate of convergence of deterministic and randomized RAS matrix scaling algorithms. Oper Res Lett 14(5):237–244
Cuturi M (2013) Sinkhorn distances: lightspeed computation of optimal transport. Adv Neural Inf Process Syst 26
Shalev-Shwartz S, Ben-David S (2014) Understanding machine learning: from theory to algorithms. Cambridge University Press, Cambridgeshire
Jones DR, Perttunen CD, Stuckman BE (1993) Lipschitzian optimization without the Lipschitz constant. J Optim Theory Appl 79(1):157–181
Peyré G, Cuturi M et al (2019) Computational optimal transport: with applications to data science. Found Trends ® Mach Learn 11(5–6):355–607
Chambolle A (2004) An algorithm for total variation minimization and applications. J Math Imaging Vision 20(1):89–97
Bartlett PL, Mendelson S (2002) Rademacher and Gaussian complexities: risk bounds and structural results. J Mach Learn Res 3(Nov):463–482
Lehmussola A, Ruusuvuori P, Selinummi J, Huttunen H, Yli-Harja O (2007) Computational framework for simulating fluorescence microscope images with cell populations. IEEE Trans Med Imaging 26(7):1010–1016
Kainz P, Urschler M, Schulter S, Wohlhart P, Lepetit V (2015) You should use regression to detect cells. In International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer pp 276–283
Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N et al (2013) The genotype-tissue expression (GTEx) project. Nature Genetics 45(6):580–585
Funding
This work is supported by the National Natural Science Foundation of China (81871508, 61773246), the Major Program of Shandong Province Natural Science Foundation (ZR2018ZB0419), the China Postdoctoral Science Foundation (No. 2021M691982), the Taishan Scholar Program of Shandong Province of China (TSHW201502038), and its 2nd round of support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, Y., Zheng, Y., Han, Z. et al. Using optimal transport theory to optimize a deep convolutional neural network microscopic cell counting method. Med Biol Eng Comput 61, 2939–2950 (2023). https://doi.org/10.1007/s11517-023-02862-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02862-7