Abstract
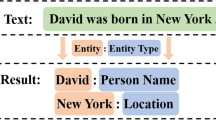
Named entity recognition (NER) is an important task in natural language processing (NLP). In recent years, NER has attracted much attention in the biomedical field. However, due to the lack of biomedical named entity identification datasets, the complexity and rarity of biomedical named entities and so on, biomedical NER is more difficult than general domain NER. So in this paper, we propose a framework (MMBERT) based on Transformer to solve the problems above. To address the scarcity of biomedical named entity recognition datasets, we introduce ERNIE-Health, a new Chinese language representation model pre-trained on large-scale biomedical text corpora. Because of the complexity and rarity of biomedical named entities, we use the Bert and CW-LSTM structures to get the joint feature vector of word pairs relations. In addition, we design multi-granularity 2D convolution to refine the relationship and representation between word pairs. Finally, we design a convolutional neural network (CNN) structure and a co-predictor to improve the model’s generalization capability and prediction accuracy. We have conducted extensive experiments on three benchmark datasets, and the experimental results show that our model achieves the best results compared with several baseline models in the experiment.
Graphical abstract






Similar content being viewed by others
Data availability
The data used in the experiment are all open data sets on the network, which meet the requirements of laws and regulations.
Code availability
The code is available at https://github.com/mmBert-Lei/MMBERT.
References
He H, Sun X (2017) F-score driven max margin neural network for named entity recognition in Chinese social media. EACL 15:713–718
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. CVPR 15:770–778
Hou F, Wang R, He J, Zhou Y (2020) Improving entity linking through semantic reinforced entity embeddings. ACL 1:6843–6848
Krauthammer M, Rzhetsky A, Morozov P et al (2000) Using BLAST for identifying gene and protein names in journal articles. Gene 259:245–252
Leaman R, Gonzalez G (2008) BANNER: an executable survey of advances in biomedical named entity recognition. Pacific Symposium Biocomputing 13:652–663
Li Y, Lin H, Yang Z (2009) Incorporating rich background knowledge for gene named entity classification and recognition.BMC Bioinforma, 10:1–10
Huang Z, Wei X, Kai Y (2015) Bidirectional LSTM-CRF models for sequence tagging. EACL 15:71–718
Zhang Y, Yang J (2018) Chinese NER Using Lattice LSTM. ACL 56:1554–1564
Vaswani A, Shazeer N, Parmar N et al (2017) Attention is all you need. Advances in Neural Information Processing Systems 30(31):5998–6008
Devlin J, Chang M W, Lee K, et al (2019) BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 conference of the north American chapter of the association for computational linguistics: human language technologies, 1:4171–4186
Ziniu W, Meng J, Jianling G et al (2019) Chinese named entity recognition method based on BERT. Comput Sci 46(S2):138–142
Collobert R, Weston J, Bottou L et al (2011) Natural language processing (almost) from scratch. J Mach Learn Res 12:2493–2537
Ma X, Hovy E (2016) End-to-end sequence labeling via Bi-directional LSTM-CNNs-CRF. In: Proceedings of the 54th annual meeting of the association for computational linguistics, vol 1, pp 1064–1074
Chiu JP, Nichols E (2016) Named entity recognition with bidirectional LSTM-CNNs. Transactions of the Association for Computational Linguistics 4:357–370
Rei M, Crichton G, Pyysalo S (2016) Attending to characters in neural sequence labeling models. In: Proceedings of COLING 2016, the 26th international conference on computational linguistics: technical papers, vol 1, pp 309–318
Tomas M, Kai C, Greg Corrado et al (2013) Efficient estimation of word representations in vector space. arXiv:1301.3781
Graziella D, Gianvito P, Michelangelo C (2022) PRILJ: an efficient two-step method based on embedding and clustering for the identification of regularities in legal case judgments. CVPR 30:359-390
Antonio P, Gianvito P, Michelangelo C (2023) SAIRUS: spatially-aware identification of risky users in social networks. Information Fusion 92:435–449
Le Q, Mikolov T (2014) Distributed representations of sentences and documents. International Conference on Machine Learning 32:1188–1196
Jenish D, Rupa M, Dipti R (2022) Effective and scalable legal judgment recommendation using pre-learned word embedding. Complex & Intelligent Systems 8(8):3199–3213
Yiming C, Wanxiang C, Ting L, et al (2020) Revisiting pre-trained models for chinese natural language processing. In: Findings of the association for computational linguistics: EMNLP 2020, vol 1, pp 657–668
Li X, Yan H, Qiu X, et al (2020) FLAT: Chinese NER using flat-lattice transformer. In: Proceedings of the 58th annual meeting of the association for computational linguistics, vol 1, pp 6836–6842
Burr S (2004) Biomedical named entity recognition using conditional random fields and rich feature set, 107-110. In: Proceedings of association for computational linguistics. Barcelona, Spain
Clark C, Aberdeen J, Coarr M et al (2010) MITRE system for clinical assertion status classification. J Am Med Inform Assoc 18(5):563–567
Xu K, Zhou Z, Hao T et al (2017) A bidirectional LSTM and conditional random fields approach to medical named entity recognition. Proceedings of international conference on advanced intelligent systems and informatics 639:355–365
Gligic L, Kormilitzin A, Goldberg P et al (2020) Named entity recognition in electronic health records using transfer learning bootstrapped neural networks. Neural Netw 121:132–139
Wang Y, Liu Y, Yu Z, et al (2012) A preliminary work on symptom name recognition from free-text clinical records of traditional Chinese medicine using conditional random fields and reasonable features, pp 223–230. Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, Montreal, Canada
Liu K, Hu Q, Liu J (2017) Named entity recognition in Chinese electronic medical records based on CRF, pp 107-110. In: Proceedings of 14th web information systems and applications conference (WISA 2017), Guangxi, China
Ya S, Jie L, Yalou H (2016) Entity recognition research in online medical texts. Journal of Peking University (Natural Science Edition) 52(1):1–9
Fan Z, Min W (2017) Medical text entities recognition method base on deep learning. Computing Technology and Automation 36(1):123–127
Chen P, Zhang M, Xiaosheng Y et al (2022) Named entity recognition of Chinese electronic medical records based on a hybrid neural network and medical MC-BERT. BMC Medical Informatics and Decision Making 22:315
Wenming Y, Weijie C (2019) Named entity recognition of online medical question answering text. Comput Syst 28(2):8–14
Tang B, Wang X, Yan J (2019) Entity recognition in Chinese clinical text using attention-based CNN-LSTM-CRF. BMC Medical Informatics and Decision Making 19(3):74–82
Cuiran P, Qinghua W, Buzhou T et al (2019) Chinese electronic medical record named entity recognition based on sentence-level Lattice-lona short-term memory neural network. Journal of the Second Military Medical University 40(5):497–506
Bo L, Xiaodong K, Huali Z et al (2020) Named entity recognition in Chinese electronic medical records using transformer-CRF. Computer Engineering and Applications 56(5):153–159
Ling L, Zhihao Y, Yawen S et al (2020) Chinese clinical named entity recognition based on stroke ELMo and multi-task learning. J Comput 43(10):1943–1957
Guoqiang T, Daqi G, Tong R et al (2020) Clinical electronic medical record named entity recognition incorporating language model. Comput Sci 47(3):211–216
Zhoufeng S, Qianmin S, Jinglei G (2021) Named entity recognition model of Chinese clinical electronic medical record based on XLNet-BiLSTM. Intelligent Computer and Applications 11(8):97–102
Qingxia Z, Wangping X, Jianqiang D et al (2021) Electronic medical record named entity recognition combined with self-attention BiLSTM-CRF. Computer Applications and Software 38(3):159–162
Zhu Y, Zhang L, Wang Y (2021) Named entity recognition on Chinese electronic medical records based on RoBERTa-WWM. Computer and Modernization 2:51–55
He T, Chen J, Wen Y (2022) Research on entity recognition of electronic medical record based on BERT-CRF Model. Computer and Digital Engineering 50(3):639–643
Jingye L, Hao F, Jiang L, et al (2022) Unified named entity recognition as word-word relation classification. In: Proceedings of the AAAI 2022 conference on artificial intelligence, 36(1):1–9
Li X, Yan H, Qiu X et al (2020) FLAT: Chinese NER using flat-lattice transformer. ACL 36(10):10965–109721
Ningyu Z, Shumin D, Zhen B, et al (2021) ERNIE-Health: a pre-trained language model for Chinese biomedical text understanding, arXiv:2110.07244
Ilias C, Manos F, Prodromos M, et al (2020) LEGAL-BERT: the muppets straight out of law school. In: Findings of the association for computational linguistics: EMNLP 2020 findings-emnlp, vol 261, pp 2898–2904
Liu Z, Mao H, Wu CY et al (2022) A ConvNet for the 2020s. CVPR 35(1):1–14
Funding
This research was supported by the National Natural Science Foundation of China(62205168), Natural Science Foundation of Fujian Province (2022J011166), Natural Science Foundation of Fujian Province(2020J01916), Natural Science Foundation of Fujian Province (2020J05109), Science and Technology Program of Putian, China(2023SZ3001PTXY13).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare no competing of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fu, L., Weng, Z., Zhang, J. et al. MMBERT: a unified framework for biomedical named entity recognition. Med Biol Eng Comput 62, 327–341 (2024). https://doi.org/10.1007/s11517-023-02934-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02934-8