Abstract
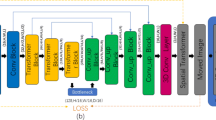
Medical image registration is a critical preprocessing step in medical image analysis. While traditional medical image registration techniques have matured, their registration speed and accuracy still fall short of clinical requirements. In this paper, we propose an improved VoxelMorph network incorporating ResNet modules and CBAM (RCV-Net), for 3D multimodal unsupervised registration. Unlike popular convolution-based U-shaped registration networks like VoxelMorph, RCV-Net incorporates the convolutional block attention module (CBAM) during the convolution process. This inclusion enhances the feature map information extraction capabilities during training and effectively prevents information loss. Additionally, we introduce a lightweight and residual network module at the network’s base, which enhances learning ability without significantly increasing training parameters. To evaluate the superiority of our registration model, we utilize evaluation metrics such as structural similarity (SSIM), peak signal-to-noise ratio (PSNR), and mean square error (MSE). Experimental results demonstrate that our proposed network structure outperforms current state-of-the-art methods, yielding better performance in multimodal registration tasks. Furthermore, generalization testing on databases outside of the training set has confirmed the optimal registration effectiveness of our model.
Graphical Abstract












Similar content being viewed by others
Data availability
The Brats 2019 dataset used during the study is a publicly available dataset from the Multimodal Brain Tumour Segmentation Challenge 2019, [https://grand-challenge.org/challenges/].
The Brats 2021 dataset used during the study is a publicly available dataset from the Multimodal Brain Tumour Segmentation Challenge 2021, [https://grand-challenge.org/challenges/].
References
Abbasi S, Tavakoli M, Boveiri HR et al (2022) Medical image registration using unsupervised deep neural network: a scoping literature review. Biomed Signal Process Control 73:103444. https://doi.org/10.1016/j.bspc.2021.103444
Bharati S, Mondal M, Podder P et al (2022) Deep learning for medical image registration: a comprehensive review. Int J Comput Inf Syst Ind Manag Appl 14:173–190. https://doi.org/10.48550/arXiv.2204.11341
Morel J-M, Yu G (2009) ASIFT: A new framework for fully affine invariant image comparison. SIAM J Imag Sci 2(2):438–469. https://doi.org/10.1137/080732730
Bay H, Tuytelaars T, Van Gool L (2006) Surf: speeded up robust features. Lect Notes Comput Sci 3951:404–417. https://doi.org/10.1007/11744023_32
Sengupta D, Gupta P, Biswas A (2022) A survey on mutual information based medical image registration algorithms. Neurocomputing 486:174–188. https://doi.org/10.1016/j.neucom.2021.11.023
Endo M, Tsunoo T, Nakamori N et al (2001) Effect of scattered radiation on image noise in cone beam CT. Med Phys 28(4):469–474. https://doi.org/10.1118/1.1357457
Avants BB, Epstein CL, Grossman M et al (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41. https://doi.org/10.1016/j.media.2007.06.004
Renner R, Wolf S (2004) Smooth Rényi entropy and applications. In: Proceedings of the 2004 IEEE International Sympsoium on Information Theory (ISIT 2004), p 233. https://doi.org/10.1109/ISIT.2004.1365269
Anastasiadis A (2012) Tsallis entropy. Entropy 14(2):174–176. https://doi.org/10.3390/e14020174
Brochet T, Lapuyade-Lahorgue J, Bougleux S et al (2021) Deep learning using havrda-charvat entropy for classification of pulmonary optical endomicroscopy. IRBM 42(6):400–406. https://doi.org/10.1016/j.irbm.2021.06.006
Studholme C, Drapaca C, Iordanova B et al (2006) Deformation-based mapping of volume change from serial brain MRI in the presence of local tissue contrast change. IEEE Trans Med Imaging 25(5):626–639. https://doi.org/10.1109/TMI.2006.872745
Sundar H, Shen D, Biros G et al (2007) Robust computation of mutual information using spatially adaptive meshes. In: International Conference on Medical Image Computing and Computer-Assisted Intervention 10(Pt 1):950–958. https://doi.org/10.1007/978-3-540-75757-3_115
Boveiri HR, Khayami R, Javidan R et al (2020) Medical image registration using deep neural networks: a comprehensive review. Comput Electr Eng 87:106767. https://doi.org/10.1016/j.compeleceng.2020.106767
Blendowski M, Heinrich MP (2019) Combining MRF-based deformable registration and deep binary 3D-CNN descriptors for large lung motion estimation in COPD patients. Int J Comput Assist Radiol Surg 14(1):43–52. https://doi.org/10.1007/s11548-018-1888-2
Eppenhof KA, Pluim JP (2018) Error estimation of deformable image registration of pulmonary CT scans using convolutional neural networks. J Med Imaging 5(2):024003. https://doi.org/10.1117/1.JMI.5.2.024003
Cheng X, Zhang L, Zheng Y (2018) Deep similarity learning for multimodal medical images. Comput Methods Biomech Biomed Eng: Imaging Vis 6(3):248–52. https://doi.org/10.1080/21681163.2015.1135299
Miao S, Wang ZJ, Liao R (2016) A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging 35(5):1352–1363. https://doi.org/10.1109/TMI.2016.2521800
Sentker T, Madesta F, Werner R (2018) GDL-FIRE: Deep learning-based fast 4D CT image registration. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing, Cham, pp 765–773. https://doi.org/10.1007/978-3-030-00928-1_86
Yan P, Xu S, Rastinehad AR et al (2018) Adversarial image registration with application for MR and TRUS image fusion. In: Machine Learning in Medical Imaging: 9th International Workshop, MLMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 16, 2018, Proceedings 9. Springer International Publishing, pp 197–204. https://doi.org/10.1007/978-3-030-00919-9_23
Lee MCH, Oktay O, Schuh A et al (2019) Image-and-spatial transformer networks for structure-guided image registration. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, Proceedings, Part II 22. Springer International Publishing, pp 337–345. https://doi.org/10.1007/978-3-030-32245-8_38
De Vos BD, Berendsen FF, Viergever MA et al (2017) End-to-end unsupervised deformable image registration with a convolutional neural network. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings 3. Springer International Publishing, pp 204–212. https://doi.org/10.1007/978-3-319-67558-9_24
Tang K, Li Z, Tian L et al (2020) ADMIR–affine and deformable medical image registration for drug-addicted brain images. IEEE Access 8:70960–70968. https://doi.org/10.1109/ACCESS.2020.2986829
Balakrishnan G, Zhao A, Sabuncu MR et al (2019) Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38(8):1788–1800. https://doi.org/10.1109/TMI.2019.2897538
Dalca AV, Balakrishnan G, Guttag J et al (2019) Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Med Image Anal 57:226–236. https://doi.org/10.1016/j.media.2019.07.006
Zhang X, Jian W, Chen Y, et al. Deform-GAN: an unsupervised learning model for deformable registration . arXiv preprint arXiv:200211430, 2020. https://doi.org/10.48550/arXiv.2002.11430
Woo S, Park J, Lee J-Y et al (2018) Cbam: convolutional block attention module. In: Computer Vision–ECCV 2018: 15th European Conference, Munich, Germany, September 8–14, 2018, Proceedings, Part VII 15. Springer International Publishing, pp 3–19. https://doi.org/10.1007/978-3-030-01234-2_1
Li X, Luo G, Wang K (2020) Multi-step cascaded networks for brain tumor segmentation. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 5th International Workshop, BrainLes 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 17, 2019, Revised Selected Papers, Part I 5. Springer International Publishing, pp 163–173. https://doi.org/10.1007/978-3-030-46640-4_16
Willmott CJ, Matsuura K (2005) Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Climate Res 30(1):79–82. https://doi.org/10.3354/cr030079
Baid U, Ghodasara S, Mohan S, et al (2021) The RSNA-ASNR-MICCAI BraTS 2021 benchmark on brain tumor segmentation and radiogenomic classification . arXiv preprint arXiv:210702314. https://doi.org/10.48550/arXiv.2107.02314
Funding
The work was supported by the National Science Foundation for Young Scientists of China (Grant No.61806060), 2019–2022, and the Natural Science Foundation of Heilongjiang Province (LH2019F024), China, 2019–2022; and Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515220140) and the Youth Innovation Project of Sun Yat-sen University Cancer Center (QNYCPY32).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by LD, QL, QZ, SH, XY, and JW The first draft of the manuscript was written by LD, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, L., Lan, Q., Zhi, Q. et al. Deep learning-based 3D brain multimodal medical image registration. Med Biol Eng Comput 62, 505–519 (2024). https://doi.org/10.1007/s11517-023-02941-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-02941-9