Abstract
In recent years, predicting gene mutations on whole slide imaging (WSI) has gained prominence. The primary challenge is extracting global information and achieving unbiased semantic aggregation. To address this challenge, we propose a novel Transformer-based aggregation model, employing a self-learning weight aggregation mechanism to mitigate semantic bias caused by the abundance of features in WSI. Additionally, we adopt a random patch training method, which enhances model learning richness by randomly extracting feature vectors from WSI, thus addressing the issue of limited data. To demonstrate the model’s effectiveness in predicting gene mutations, we leverage the lung adenocarcinoma dataset from Shandong Provincial Hospital for prior knowledge learning. Subsequently, we assess TP53, CSMD3, LRP1B, and TTN gene mutations using lung adenocarcinoma tissue pathology images and clinical data from The Cancer Genome Atlas (TCGA). The results indicate a notable increase in the AUC (Area Under the ROC Curve) value, averaging 4%, attesting to the model’s performance improvement. Our research offers an efficient model to explore the correlation between pathological image features and molecular characteristics in lung adenocarcinoma patients. This model introduces a novel approach to clinical genetic testing, expected to enhance the efficiency of identifying molecular features and genetic testing in lung adenocarcinoma patients, ultimately providing more accurate and reliable results for related studies.
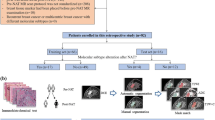
Graphical abstract
Novel aggregation model based on transformer. The WSI selects tumor patches via the tumor classification model and then inputs them into the Swin Transformer backbone network to extract tumor vectors. The extracted vectors are trained using the random patch training method, and then processed through a transformer encoder and weighted pooling layer to converge into a feature vector that represents global information. The predicted results are then outputted.










Similar content being viewed by others
References
Hayashi R, Inomata M (2022) Small cell lung cancer; recent advances of its biology and therapeutic perspective. Respiratory Invest 60(2):197–204
Wu J, Lin Z (2022) Non-small cell lung cancer targeted therapy: Drugs and mechanisms of drug resistance. Int J Molecular Sci 23(23):15056
Bourbonne V, Geier M, Schick U, Lucia F (2022) Multi-omics approaches for the prediction of clinical endpoints after immunotherapy in non-small cell lung cancer: A comprehensive review. Biomedicines 10(6):1237
Aalami AH, Aalami F (2022) Diagnostic performance of fluorescence in situ hybridization (fish) in upper tract urothelial carcinoma (utuc): a systematic review and meta-analysis. Int J Clinical Oncol 27(10):1605–1615
Pervez MT, Abbas SH, Moustafa MF, Aslam N, Shah SSM et al. (2022) A comprehensive review of performance of next-generation sequencing platforms. BioMed Res Int 2022
Nassif AB, Talib MA, Nasir Q, Afadar Y, Elgendy O (2022) Breast cancer detection using artificial intelligence techniques: A systematic literature review. Artif Intell Med 102276
Runa F, Hamalian S, Meade K, Shisgal P, Gray P, Kelber J (2017) Tumor microenvironment heterogeneity: challenges and opportunities. Curr Molecul Biol Reports 3:218–229
Arneth B (2019) Tumor microenvironment. Medicina 56(1):15
Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Kolahian S, Javaheri T, Zare P (2020) Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal 18:1–19
Abousamra S, Gupta R, Hou L, Batiste R, Zhao T, Shankar A, Rao A, Chen C, Samaras D, Kurc T et al (2022) Deep learning-based mapping of tumor infiltrating lymphocytes in whole slide images of 23 types of cancer. Front Oncol 11:5971
Baxi V, Edwards R, Montalto M, Saha S (2022) Digital pathology and artificial intelligence in translational medicine and clinical practice. Modern Pathol 35(1):23–32
Wang X, Xie T, Luo J, Zhou Z, Yu X, Guo X (2022) Radiomics predicts the prognosis of patients with locally advanced breast cancer by reflecting the heterogeneity of tumor cells and the tumor microenvironment. Breast Cancer Res 24(1):20
Di D, Zhang J, Lei F, Tian Q, Gao Y (2022) Big-hypergraph factorization neural network for survival prediction from whole slide image. IEEE Trans. Image Process 31:1149–1160
Arunachalam HB, Mishra R, Daescu O, Cederberg K, Rakheja D, Sengupta A, Leonard D, Hallac R, Leavey P (2019) Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PloS one 14(4):0210706
Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, Moreira AL, Razavian N, Tsirigos A (2018) Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nature Med 24(10):1559–1567
Tomita N, Tafe LJ, Suriawinata AA, Tsongalis GJ, Nasir-Moin M, Dragnev K, Hassanpour S (2022) Predicting oncogene mutations of lung cancer using deep learning and histopathologic features on whole-slide images. Translational Oncol 24:101494
He K, Gan C, Li Z, Rekik I, Yin Z, Ji W, Gao Y, Wang Q, Zhang J, Shen D (2022) Transformers in medical image analysis: A review. Intell Med
Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, Fu H (2022) Transformers in medical imaging: A survey. arXiv:2201.09873
Ma Y, Jiang Z, Zhang H, Xie F, Zheng Y, Shi H, Zhao Y (2016) Breast histopathological image retrieval based on latent dirichlet allocation. IEEE J Biomed Health Inf 21(4):1114–1123
Veta M, Heng YJ, Stathonikos N, Bejnordi BE, Beca F, Wollmann T, Rohr K, Shah MA, Wang D, Rousson M et al (2019) Predicting breast tumor proliferation from whole-slide images: the tupac16 challenge. Med Image Anal 54:111–121
Ukwuoma CC, Hossain MA, Jackson JK, Nneji GU, Monday HN, Qin Z (2022) Multi-classification of breast cancer lesions in histopathological images using deep_pachi: Multiple self-attention head. Diagnostics 12(5):1152
Shao Z, Bian H, Chen Y, Wang Y, Zhang J, Ji X et al (2021) Transmil: Transformer based correlated multiple instance learning for whole slide image classification. Adv Neural Inf Process Syst 34:2136–2147
Xu H, Park S, Hwang TH (2019) Computerized classification of rostate cancer gleason scores from whole slide images. IEEE/ACM Trans Comput Biol Bioinform 17(6):1871–1882
Ding M, Qu A, Zhong H, Liang H (2021) A transformer-based network for pathology image classification. In: 2021 IEEE International conference on bioinformatics and biomedicine (BIBM), IEEE, pp 2028–2034
Wang S, Yang DM, Rong R, Zhan X, Fujimoto J, Liu H, Minna J, Wistuba II, Xie Y, Xiao G (2019) Artificial intelligence in lung cancer pathology image analysis. Cancers 11(11):1673
Schneider L, Laiouar-Pedari S, Kuntz S, Krieghoff-Henning E, Hekler A, Kather JN, Gaiser T, Fröhling S, Brinker TJ (2022) Integration of deep learning-based image analysis and genomic data in cancer pathology: a systematic review. Eur J Cancer 160:80–91
Linkon AHM, Labib MM, Hasan T, Hossain M et al (2021) Deep learning in prostate cancer diagnosis and gleason grading in histopathology images: An extensive study. Inf Med Unlocked 24:100582
Schmauch B, Romagnoni A, Pronier E, Saillard C, Maillé P, Calderaro J, Kamoun A, Sefta M, Toldo S, Zaslavskiy M et al. A deep learning model to predict rna-seq expression of tumours from whole slide images. Nature Commun 11(1):3877
Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N (2021) Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med 13(1):1–17
Di D, Li S, Zhang J, Gao Y (2020) Ranking-based survival prediction on histopathological whole-slide images. In: Medical image computing and computer assisted intervention–MICCAI 2020: 23rd international conference, Lima, Peru, October 4–8, 2020, Proceedings, Part V 23, Springer, pp 428–438
Qu H, Zhou M, Yan Z, Wang H, Rustgi VK, Zhang S, Gevaert O, Metaxas DN (2021) Genetic mutation and biological pathway prediction based on whole slide images in breast carcinoma using deep learning. NPJ Precision Oncol 5(1):87
Chen Z, Wang M, De Wilde RL, Feng R, Su M, Torres-de la Roche LA, Shi W (2021) A machine learning model to predict the triple negative breast cancer immune subtype. Front Immunol 12:749459
Funding
This work is supported by the National Nature Science Foundation of China (No.: 62272283), the Major Basic Research Project of Shandong Natural Science Foundation (No.: ZR2019ZD04) and the New Twentieth Items of Universities in Jinan (2021GXRC049).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, K., Zheng, Y., Yang, X. et al. A novel transformer-based aggregation model for predicting gene mutations in lung adenocarcinoma. Med Biol Eng Comput 62, 1427–1440 (2024). https://doi.org/10.1007/s11517-023-03004-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-023-03004-9