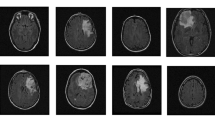
Abstract
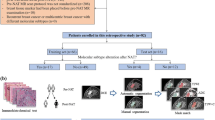
Invasive gene expression profiling studies have exposed prognostically significant breast cancer subtypes: normal-like, luminal, HER-2 enriched, and basal-like, which is defined in large part by human epidermal growth factor receptor 2 (HER-2), progesterone receptor (PR), and estrogen receptor (ER). However, while dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) has been generally employed in the screening and therapy of breast cancer, there is a challenging problem to noninvasively predict breast cancer molecular subtypes, which have extremely low-data regimes. In this paper, a novel few-shot learning scheme, which combines lightweight contrastive convolutional neural network (LC-CNN) and multi-contrast learning strategy (MCLS), is worthwhile to be developed for predicting molecular subtype of breast cancer in DCE-MRI. Moreover, MCLS is designed to construct One-vs-Rest and One-vs-One classification tasks, which addresses inter-class similarity among normal-like, luminal, HER-2 enriched, and basal-like. Extensive experiments demonstrate the superiority of our proposed scheme over state-of-the-art methods. Furthermore, our scheme is able to achieve competitive results on few samples due to joint LC-CNN and MCLS for excavating contrastive correlations of a pair of DCE-MRI.
Graphical Abstract









Similar content being viewed by others
Code availability
Our source code will be released on https://github.com/AI-medical-diagnosis-team-of-JNU/LCCNN-Classification.
References
Shu X, Zhang L, Wang Z et al (2020) Deep neural networks with region-based pooling structures for mammographic image classification. IEEE Trans Med Imaging 39:2246–2255. https://doi.org/10.1109/TMI.2020.2968397
Zhou Y, Chen H, Li Y et al (2021) Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Med Image Anal 70:101918. https://doi.org/10.1016/j.media.2020.101918
Bertucci F, Finetti P, Rougemont J et al (2005) Gene expression profiling identifies molecular subtypes of inflammatory breast cancer. Can Res 65:2170–2178. https://doi.org/10.1158/0008-5472.CAN-04-4115
Uddin MdN, Wang X (2022) Identification of breast cancer subtypes based on gene expression profiles in breast cancer stroma. Clin Breast Cancer 22:521–537. https://doi.org/10.1016/j.clbc.2022.04.001
Zhang Y, Chen J-H, Lin Y et al (2021) Prediction of breast cancer molecular subtypes on DCE-MRI using convolutional neural network with transfer learning between two centers. Eur Radiol 31:2559–2567. https://doi.org/10.1007/s00330-020-07274-x
Esposito A, Criscitiello C, Locatelli M et al (2016) Liquid biopsies for solid tumors: understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol Ther 157:120–124. https://doi.org/10.1016/j.pharmthera.2015.11.007
Huang H, Li H (2021) Tumor heterogeneity and the potential role of liquid biopsy in bladder cancer. Cancer Commun 41:91–108. https://doi.org/10.1002/cac2.12129
Lee JY, Lee K, Seo BK et al (2022) Radiomic machine learning for predicting prognostic biomarkers and molecular subtypes of breast cancer using tumor heterogeneity and angiogenesis properties on MRI. Eur Radiol 32:650–660. https://doi.org/10.1007/s00330-021-08146-8
Chung M, Calabrese E, Mongan J et al (2023) Deep learning to simulate contrast-enhanced breast MRI of invasive breast cancer. Radiology 306:e213199. https://doi.org/10.1148/radiol.213199
Wang W, Lv S, Xun J et al (2022) Comparison of diffusion kurtosis imaging and dynamic contrast enhanced MRI in prediction of prognostic factors and molecular subtypes in patients with breast cancer. Eur J Radiol 154:110392. https://doi.org/10.1016/j.ejrad.2022.110392
Krizhevsky A, Sutskever I, Hinton GE (2017) ImageNet classification with deep convolutional neural networks. Commun ACM 60:84–90. https://doi.org/10.1145/3065386
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. https://doi.org/10.48550/arXiv.1409.1556
Lecun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86:2278–2324. https://doi.org/10.1109/5.726791
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, Las Vegas, NV, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, van der Maaten L, Weinberger KQ (2018) Densely connected convolutional networks. https://doi.org/10.48550/arXiv.1608.06993
Roth HR, Lee CT, Shin H-C et al (2015) Anatomy-specific classification of medical images using deep convolutional nets. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI 2015). IEEE, Brooklyn, NY, pp 101–104. https://doi.org/10.1109/ISBI.2015.7163826
Pawlowski N, Ktena SI, Lee MCH et al (2017) DLTK: state of the art reference implementations for deep learning on medical images. https://doi.org/10.48550/arXiv.1711.06853
Li G, Li C, Wu G et al (2022) MF-OMKT: model fusion based on online mutual knowledge transfer for breast cancer histopathological image classification. Artif Intell Med 134:102433. https://doi.org/10.1016/j.artmed.2022.102433
Teng J, Zhang H, Liu W et al (2022) A dynamic Bayesian model for breast cancer survival prediction. IEEE J Biomed Health Inform 26:5716–5727. https://doi.org/10.1109/JBHI.2022.3202937
Abbet C, Studer L, Fischer A et al (2022) Self-rule to multi-adapt: generalized multi-source feature learning using unsupervised domain adaptation for colorectal cancer tissue detection. Med Image Anal 79:102473. https://doi.org/10.1016/j.media.2022.102473
Jiang Y, Xu S, Fan H et al (2021) ALA-Net: adaptive lesion-aware attention network for 3D colorectal tumor segmentation. IEEE Trans Med Imaging 40:3627–3640. https://doi.org/10.1109/TMI.2021.3093982
Yoo TK, Choi JY, Kim HK (2021) Feasibility study to improve deep learning in OCT diagnosis of rare retinal diseases with few-shot classification. Med Biol Eng Comput 59:401–415. https://doi.org/10.1007/s11517-021-02321-1
Vanschoren J (2018) Meta-learning: a survey. https://doi.org/10.48550/arXiv.1810.03548
Snell J, Swersky K, Zemel RS (2017) Prototypical networks for few-shot learning. https://doi.org/10.48550/arXiv.1703.05175
Vinyals O, Blundell C, Lillicrap T et al (2017) Matching networks for one shot learning. https://doi.org/10.48550/arXiv.1606.04080
Finn C, Abbeel P, Levine S (2017) Model-agnostic meta-learning for fast adaptation of deep networks. https://doi.org/10.48550/arXiv.1703.03400
Han K, Wang Y, Tian Q et al (2020) GhostNet: more features from cheap operations. https://doi.org/10.48550/arXiv.1911.11907
Guo S, Lai B, Yang S et al (2023) Sensitivity pruner: filter-level compression algorithm for deep neural networks. Pattern Recogn 140:109508. https://doi.org/10.1016/j.patcog.2023.109508
Young S, Wang Z, Taubman D, Girod B (2021) Transform quantization for CNN compression. IEEE Trans Pattern Anal Mach Intell 1–1. https://doi.org/10.1109/TPAMI.2021.3084839
Nekooei A, Safari S (2022) Compression of deep neural networks based on quantized tensor decomposition to implement on reconfigurable hardware platforms. Neural Netw 150:350–363. https://doi.org/10.1016/j.neunet.2022.02.024
Suzuki T, Huang L (2022) Edge-aware extended Star-Tetrix Transforms For CFA-sampled raw camera image compression. IEEE Trans on Image Process 31:6072–6082. https://doi.org/10.1109/TIP.2022.3205470
Shi R, Niu L, Zhou R (2022) Sparse CapsNet with explicit regularizer. Pattern Recogn 124:108486. https://doi.org/10.1016/j.patcog.2021.108486
Amendola G, Greco G, Veltri P (2022) Answers set programs for non-transferable utility games: expressiveness, complexity and applications. Artif Intell 302:103606. https://doi.org/10.1016/j.artint.2021.103606
Tan Y, Yang K-F, Zhao S-X, Li Y-J (2022) Retinal vessel segmentation with skeletal prior and contrastive loss. IEEE Trans Med Imaging 41:2238–2251. https://doi.org/10.1109/TMI.2022.3161681
Zhu P, Zhu Z, Wang Y et al (2022) Multi-granularity episodic contrastive learning for few-shot learning. Pattern Recogn 131:108820. https://doi.org/10.1016/j.patcog.2022.108820
Liu Z, Zhu Z, Zheng S et al (2022) Margin preserving self-paced contrastive learning towards domain adaptation for medical image segmentation. IEEE J Biomed Health Inform 26:638–647. https://doi.org/10.1109/JBHI.2022.3140853
Newitt D, Hylton N (2016) Single site breast DCE-MRI data and segmentations from patients undergoing neoadjuvant chemotherapy. The Cancer Imaging Archive 2
Cheang MCU, Chia SK, Voduc D et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal b breast cancer. JNCI: J Natl Cancer Inst 101:736–750. https://doi.org/10.1093/jnci/djp082
Koch G, Zemel R, Salakhutdinov R (2015) Siamese neural networks for one-shot image recognition. ICML deep learning workshop 2:1
Jiang H, Ma H, Qian W et al (2018) An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J Biomed Health Inform 22:1227–1237. https://doi.org/10.1109/JBHI.2017.2725903
Hadsell R, Chopra S, LeCun Y (2006) Dimensionality reduction by learning an invariant mapping. In: 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition - Volume 2 (CVPR’06). IEEE, New York, NY, pp 1735–1742. https://doi.org/10.1109/CVPR.2006.100
Sung F, Yang Y, Zhang L et al (2018) Learning to compare: relation network for few-shot learning. https://doi.org/10.48550/arXiv.1711.06025
Shorfuzzaman M, Hossain MS (2021) MetaCOVID: a Siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recogn 113:107700. https://doi.org/10.1016/j.patcog.2020.107700
Cai A, Hu W, Zheng J (2020) Few-shot learning for medical image classification. Artificial Neural Networks and Machine Learning – ICANN 2020. Springer International Publishing, Cham, pp 441–452
Li X, Yu L, Jin Y et al (2020) Difficulty-aware meta-learning for rare disease diagnosis. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. Springer International Publishing, Cham, pp 357–366
Leung CK, Madill EWR, Tran NDT, Zhang CY (2022) Health informatics on big COVID-19 pandemic data via n-shot learning. In: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, Las Vegas, NV, pp 2784–2791. https://doi.org/10.1109/BIBM55620.2022.9995592
Zhang K, Qi S, Cai J et al (2022) Content-based image retrieval with a convolutional Siamese neural network: distinguishing lung cancer and tuberculosis in CT images. Comput Biol Med 140:105096. https://doi.org/10.1016/j.compbiomed.2021.105096
Funding
This work is supported in part by the National Key R&D Program of China under Grants 2021YFE0203700 and 2018YFA0701700; the Postgraduate Research & Practice Innovation Program of Jiangsu Province SJCX22_1106; the National Natural Science Foundation of China Grants 61602007, U21A20521, and 61731008; the Zhejiang Provincial Natural Science Foundation of China (LZ15F010001), the Jiangsu Provincial Maternal and Child Health Research Project (F202034), the Wuxi Health Commission Precision Medicine Project (J202106); the Jiangsu Provincial Six Talent Peaks Project (YY-124), and the Science and Technology Development Fund, Macau SAR (File no. 0004/2019/AFJ and 0011/2019/AKP).
Author information
Authors and Affiliations
Contributions
Chunjuan Jiang and Yan Zhang are the guarantors of this study. Xiang Pan, Yihang Wang, Yuan Liu, and Yan Zhang collected the data and performed the preprocessing. Xiang Pan, Yihang Wang, and Pei Wang designed and conducted the experiments. Xiang Pan, Pei Wang, Yihang Wang, and Chunjuan Jiang performed statistical analyses of the results. Xiang Pan, Pei Wang, Shunyuan Jia, and Yihang Wang drafted the manuscript. Pei Wang, Yan Zhang, and Shunyuan Jia revised the paper. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Ethical approval
No human or animal subjects were involved in this study. Hence, no ethical approval is required.
Consent for publication
All authors involved consent to publish work here within.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, X., Wang, P., Jia, S. et al. Multi-contrast learning-guided lightweight few-shot learning scheme for predicting breast cancer molecular subtypes. Med Biol Eng Comput 62, 1601–1613 (2024). https://doi.org/10.1007/s11517-024-03031-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-024-03031-0