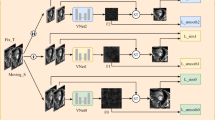
Abstract
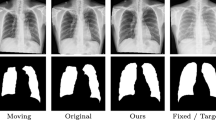
Image registration is a primary task in various medical image analysis applications. However, cardiac image registration is difficult due to the large non-rigid deformation of the heart and the complex anatomical structure. This paper proposes a structure-aware independently trained multi-scale registration network (SIMReg) to address this challenge. Using image pairs of different resolutions, independently train each registration network to extract image features of large deformation image pairs at different resolutions. In the testing stage, the large deformation registration is decomposed into a multi-scale registration process, and the deformation fields of different resolutions are fused by a step-by-step deformation method, thus solving the difficulty of directly processing large deformation. Meanwhile, the targeted introduction of MIND (modality independent neighborhood descriptor) structural features to guide network training enhances the registration of cardiac structural contours and improves the registration effect of local details. Experiments were carried out on the open cardiac dataset ACDC (automated cardiac diagnosis challenge), and the average Dice value of the experimental results of the proposed method was 0.833. Comparative experiments showed that the proposed SIMReg could better solve the problem of heart image registration and achieve a better registration effect on cardiac images.
Graphical Abstract







Similar content being viewed by others
References
Khalil A, Ng S C, Liew Y M, et al (2018) An overview on image registration techniques for cardiac diagnosis and treatment. Cardiol Res Pract. https://doi.org/10.1155/2018/1437125
Klein GJ, Huesman RH (2002) Four-dimensional processing of deformable cardiac PET data. Med Image Anal 6(1):29–46
Jr R, Walter J et al (1991) Quantification of and correction for left ventricular systolic long-axis shortening by magnetic resonance tissue tagging and slice isolation. Circulation 84(2):721–731
O’Dell WG, Moore CC, Hunter WC et al (1995) Three-dimensional myocardial deformations: calculation with displacement field fitting to tagged MR images. Radiology 195(3):829–835
Avendi MR, Kheradvar A, Jafarkhani H (2017) Automatic segmentation of the right ventricle from cardiac MRI using a learning-based approach. Magn Reson Med 78(6):2439–2448
Tavakoli V, Amini AA (2013) A survey of shaped-based registration and segmentation techniques for cardiac images. Comput Vis Image Underst 117(9):966–989
Zhou Y, Pang S, Cheng J et al (2020) Unsupervised deformable medical image registration via pyramidal residual deformation fields estimation. arXiv preprint arXiv:200407624. https://doi.org/10.48550/arXiv.2004.07624
Hu X, Kang M, Huang W, et al. (2019) Dual-stream pyramid registration network. Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part II. Cham: Springer International Publishing, 382-390
Zhang L, Zhou L, Li R et al (2021) Cascaded feature warping network for unsupervised medical image registration. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), vol 2021. pp 913–916. https://doi.org/10.1109/ISBI48211.2021.9433880
Dosovitskiy A, Fischer P, Ilg E et al (2015) Flownet: Learning optical flow with convolutional networks. In: Proceedings of the IEEE International Conference on Computer Vision. pp 2758–2766
Cao Y, Zhu Z, Rao Y et al (2021) Edge-aware pyramidal deformable network for unsupervised registration of brain MR images. Front Neurosci 14:620235
Fechter T, Baltas D (2020) One-shot learning for deformable medical image registration and periodic motion tracking. IEEE Trans Med Imaging 39(7):2506–2517
Yu H, et al. (2020) Motion pyramid networks for accurate and efficient cardiac motion estimation. Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part VI 23. Springer International Publishing. https://doi.org/10.1007/978-3-030-59725-2_42
Li H, Fan Y, Alzheimer’s Disease Neuroimaging Initiative (2022) MDReg-Net: multi-resolution diffeomorphic image registration using fully convolutional networks with deep self-supervision. Hum Brain Mapp 43(7):2218–2231
Mok T C W, Chung A C S (2020) Large deformation diffeomorphic image registration with Laplacian pyramid networks. Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part III 23. Springer International Publishing, 211-221
Han R, Jones CK, Lee J et al (2022) Joint synthesis and registration network for deformable MR-CBCT image registration for neurosurgical guidance. Phys Med Biol 67(12):125008
Heinrich MP, Jenkinson M, Bhushan M et al (2012) MIND: Modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal 16(7):1423–1435
Jiang Z, Yin FF, Ge Y et al (2020) A multi-scale framework with unsupervised joint training of convolutional neural networks for pulmonary deformable image registration. Phys Med Biol 65(1):015011
Eppenhof KAJ, Lafarge MW, Veta M et al (2019) Progressively trained convolutional neural networks for deformable image registration. IEEE Trans Med Imaging 39(5):1594–1604
Milletari F, Navab N, Ahmadi SA (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 2016 Fourth International Conference on 3D Vision (3DV), vol 2016. Stanford, CA, USA, pp 565–571. https://doi.org/10.1109/3DV.2016.79
Bernard O, Lalande A, Zotti C et al (2018) Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging 37(11):2514–2525
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight J 2(365):1–35
Thirion JP (1998) Image matching as a diffusion process: an analogy with Maxwell’s demons. Med Image Anal 2(3):243–260
Avants BB, Epstein CL, Grossman M et al (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Balakrishnan G, Zhao A, Sabuncu MR et al (2019) Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38(8):1788–1800
Chen P, Chen X, Chen E et al (2020) Anatomy-aware cardiac motion estimation. International workshop on machine learning in medical imaging. Springer International Publishing, Cham, pp 150–159
Zheng Z, Cao W, Duan Y et al (2022) Multi-strategy mutual learning network for deformable medical image registration. Neurocomputing 501:102–112
Krebs J, Delingette H, Mailhé B et al (2019) Learning a probabilistic model for diffeomorphic registration. IEEE Trans Med Imaging 38(9):2165–2176
Funding
This work was supported by the National Natural Science Foundation of China under award number 61976091.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Qing Chang and Yaqi Wang. The first draft of the manuscript was written by Qing Chang and Yaqi Wang, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
This work does not involve any human participant or animal.
All authors are informed.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chang, Q., Wang, Y. Structure-aware independently trained multi-scale registration network for cardiac images. Med Biol Eng Comput 62, 1795–1808 (2024). https://doi.org/10.1007/s11517-024-03039-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-024-03039-6