Abstract
Purpose
To develop a novel automated method for segmentation of the injured spleen using morphological properties following abdominal trauma. Average attenuation of a normal spleen in computed tomography (CT) does not vary significantly between subjects. However, in the case of solid organ injury, the shape and attenuation of the spleen on CT may vary depending on the time and severity of the injury. Timely assessment of the severity and extent of the injury is of vital importance in the setting of trauma.
Methods
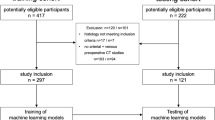
We developed an automated computer-aided method for segmenting the injured spleen from CT scans of patients who had splenectomy due to abdominal trauma. We used ten subjects to train our computer-aided diagnosis (CAD) method. To validate the CAD method, we used twenty subjects in our testing group. Probabilistic atlases of the spleens were created using manually segmented data from ten CT scans. The organ location was modeled based on the position of the spleen with respect to the left side of the spine followed by the extraction of shape features. We performed the spleen segmentation in three steps. First, we created a mask of the spleen, and then we used this mask to segment the spleen. The third and final step was the estimation of the spleen edges in the presence of an injury such as laceration or hematoma.
Results
The traumatized spleens were segmented with a high degree of agreement with the radiologist-drawn contours. The spleen quantification led to \(86\pm 5\,\%\) volume overlap, \(92.5\pm 3.11\,\%\) Dice similarity index, \(89.05\pm 5.29\,\%/96.42\pm 2.55\) precision/sensitivity, \(8\pm 5\,\%\) volume estimation error rate, \(1.09\pm 0.62/1.91\pm 1.45\,\hbox {mm}\) average surface distance/root-mean-squared error.
Conclusions
Our CAD method robustly segments the spleen in the presence of morphological changes such as laceration, contusion, pseudoaneurysm, active bleeding, periorgan and parenchymal hematoma, including subcapsular hematoma due to abdominal trauma. CAD of the splenic injury due to abdominal trauma can assist in rapid diagnosis and assessment and guide clinical management. Our segmentation method is a general framework that can be adapted to segment other injured solid abdominal organs.

















Similar content being viewed by others
References
Nishijima DK, Simel DL, Wisner DH, Holmes JF (2012) Does this adult patient have a blunt intra-abdominal injury? JAMA 307(14):1517–1527
Poletti PA, Wintermark M, Schneyder P, Becker CD (2002) Traumatic injuries: role of imaging in the management of the polytrauma victim (conservative expectation). Eur Radiol 12:969–978
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310
Erdt M, Steger S, Sakas G (2012) Regmentation: a new view of image segmentation and registration. J Radiat Oncol Inf 4(1):1–23
Li J, He B, Jiang H, Yang B, Zhang L, Tong O, Dong A (2013) A Web Abdominal Anatomical Structure Database System.In: Proceedings of ICMIPE, pp 19–23
Campadelli P, Casiraghi E, Pratissoli S (2009) Automatic abdominal organ segmentation from CT images. J. Sound and Vib 8:1–14
Ruskó L, Bekes G, Németh G, Fidrich M (2007) Fully automatic liver segmentation for contrast-enhanced CT images.In: Proceedings of MICCAI workshop 3-D segment clinic, grand challenge, pp 143–150
Kaneko T, Gu L, Fujimoto H (2002) Recognition of abdominal organs using 3D mathematical morphology. Syst Comput Jpn 33(8):75–83
Grau V, Mewes AUJ, Alcañiz M, Kikinis R, Warfield SK (2004) Improved watershed transform for medical image segmentation using prior information. IEEE Trans Med Imaging 23(4):447–458
Behrad A, Masoumi H (2010) Automatic spleen segmentation in MRI images using a combined neural network and recursive watershed transform. In Proceedings on NEUREL, pp 63–67
Linguraru MG, Pura JA, Chowdhury AS, Summers RM (2010) Multi-organ segmentation from multi-phase abdominal CT via 4D graphs using enhancement, shape and location optimization. Med Image Comput Assist Interv 13:89–96
Linguraru MG, Pura JA, Pamulapati V, Summers RM (2012) Statistical 4D graphs for multi-organ abdominal segmentation from multiphase CT. Med Image Anal 16:904–914
Boykov Y, Kolmogorov V (2004) An experimental comparison of mincut/ max-flow algorithms. IEEE Trans Pattern Anal Mach Intell 26(9):1124–1137
Kolmogorov V, Zabih R (2004) What energy functions can be minimized via graph cuts? IEEE Trans Pattern Anal Mach Intell 26(2):147–159
Veksler O (2008) Star-shape prior for graph-cut image segmentation. In: Proceedings on 10th Eur Conf Comput Vis, pp 454–467
Linguraru MG, Summers RM (2008) Multi-organ automatic segmentation in 4D contrast-enhanced abdominal CT. Biomedical imaging: from nano to macro 2008 IEEE international symposium, pp 45–48
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1(4):321–331
Liu F, Zhao B, Kijewski PK, Wang L, Schwartz LH (2005) Liver segmentation for CT images using GVF snake. Med Phys 32(12):3699–3706
McInerney T, Terzopoulos D (2000) Deformable Models, Handbook of Medical Imaging: Processing and Analysis. In: Bankman I (ed) Academic, New York
Park H, Bland PH, Meyer CR (2003) Construction of an abdominal probabilistic atlas and its application in segmentation. IEEE Trans on Med Imaging 22(4):483–492
Linguraru MG, Sandberg JK, Li ZX, Shah F, Summers RM (2010) Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med Phys 37(2):771–783
Wolz R, Chu C, Misawa K, Fujiwara M, Mori K, Rueckert D (2013) Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans on Med Imaging 32(9):1723–1730
Okada T, Linguraru MG, Hori M, Suzuki Y, Summers RM, Tomiyama N, Sato Y (2012) Multi-organ segmentation in abdominal CT images. 34th Annual International Conference of the IEEE EMBS, pp 3986–3989
Cuadra MB, Pollo C, Bardera A, Cuisenaire O, Villemure JG, Thiran JP (2004) Atlas-based segmentation of pathological MR brain images using a model of lesion growth. IEEE Trans Med Imaging 23(10):1301–1314
Basin PL, Pham DL (2008) Homeomorphic brain image segmentation with topological and statistical atlases. Med Image Anal 12(5):616–625
Noble JH, Dawant BM (2009) Automatic segmentation of the optic nerves and chiasm in CT and MR using the atlas-navigated optimal medial axis and deformable-model algorithm. In: Proceedings on SPIE 7259, pp 725 916-1–725 916-10
Zhou X, Hayashi T, Han M, Chen H, Hara T, Fujita H, Yokoyama R, Kanematsu M, Hoshi H (2009) Automated segmentation and recognition of the bone structure in non-contrast torso CT images using implicit anatomical knowledge. In Proceedings on SPIE 7259, pp 72 583-4–72 593S-1
Campadelli P, Casiraghi E, Pratissoli S (2011) A segmentation framework for abdominal organs from CT scans. Artif Intell Med 50:3–11
Campadelli P, Casiraghi E, Pratissoli S (2008) Fully automatic segmentation of abdominal organs from CT images using fast marching methods. 21st IEEE international symposium on computer-based med systems, pp 554–559
Malladi R, Sethian JA, Vemuri BC (1995) Shape modeling with front propagation: a level set approach. IEEE Trans Pattern Anal Mach Intell 17(2):158–175
Zhang X, Fujita H, Qina T, Zhaod J (2008) A novel method for extraction of spleen by using thin plate splines (TPS) deformation and edge detection from abdominal CT images. Proc BMEI 1:830–834
Gonzalo RD, Chenouard N, Unser M (2013) Spline-based deforming ellipsoids for interactive 3D bioimage segmentation. IEEE Trans on Image Process 22(10):3926–3940
Gollmer ST, Simon M, Bischof A, Barkhausen J, Buzug TM (2012) Multi-object active shape model construction for abdomen segmentation: preliminary results. 34th Annual international conference of the IEEE EMBS, pp 3990–3994
Frangi AF, Rueckert D, Schnabel JA, Niessen WJ (2002) Automatic construction of multiple-object three-dimensional statistical shape models: application to cardiac modeling. IEEE Trans Med Imaging 21(9):1151–1166
Seifert S, Barbu A, Zhou SK, Liu D, Feulner J, Huber M, Suehling M, Cavallaro A, Comaniciu D (2009) Hierarchical parsing and semantic navigation of full body CT data. In: Proceedings on SPIE 7259, id 725
Ling H, Zhou SK, Zheng Y, Georgescu B, Suehling M, Comaniciu D (2008) Hierarchical, learning-based automatic liver segmentation. In: Proceedings on IEEE computer society conference on computer vision pattern recognition, pp 1–8
Cootes TF, Taylor CJ, Cooper DH, Graham J (1995) Active shape models—their training and application. Comput Vis Image Underst 61(1):38–59
Cootes TF, Edwards G, Taylor C (2001) Active appearance models. IEEE Trans Pattern Anal Mach Intell 23(6):681–685
Stegmann MB, Ersbll BK, Larsen R (2003) FAME—a flexible appearance modeling environment. IEEE Trans Med Imaging 22(10):1319–1331
Mitchell SC, Lelieveldt BPF, van der Geest RJ, Bosch HG, Reiber JHC, Sonka M (2001) Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. IEEE Trans Med Imaging 20(5):415–423
Jiang H, Ma Z, Zhang B, Zhang Y (2011) A spleen segmentation method based on PCA-ISO. Proc ICCIT 6:928–933
Li C, Wang X, Li J, Eberl S, Fulham M, Yin Y, Feng DD (2013) Joint probabilistic model of shape and intensity for multiple abdominal organ segmentation from volumetric CT images. IEEE J Biomed Health Inf 17(1):92–102
Chen X, Udupa JK, Bagci U, Zhuge Y, Yao J (2012) Medical image segmentation by combining graph cuts and oriented active appearance models. IEEE Trans on Image Process 21(4):2035–2046
Liu X, Linguraru MG, Yao J, Summer RM (2011) Abdominal multi-organ localization on contrast-enhanced Ct based on maximum a posteriori probability and minimum volume overlap. Biomedical imaging: from nano to macro IEEE international symposium, pp 2083–2086
Jawarneh MS, Mandava R, Ramachandram D, Shuaib IL (2010) Automatic initialization of contour for level set algorithms guided by integration of multiple views to segment abdominal CT scans. In: Proceedings of CIMSiM, pp 315–320
Rezai P, Tochetto SM, Galizia MS, Yaghmai V (2011) Splenic volume model constructed from standardized one-dimensional MDCT measurements. Am J Roentgenol 196(2):367–372. doi:10.2214/AJR.10.4453
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
O. Dandin, U. Teomete, O. Osman, G. Tulum, T. Ergin, and M.Z. Sabuncuoglu declare that they have no conflicts of interest in the research.
Informed consent
This study was approved by Institutional Review Board (IRB) of University of Miami, and informed consent was waived by the IRB due to its retrospective design.
Rights and permissions
About this article
Cite this article
Dandin, O., Teomete, U., Osman, O. et al. Automated segmentation of the injured spleen. Int J CARS 11, 351–368 (2016). https://doi.org/10.1007/s11548-015-1288-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-015-1288-9