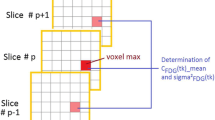
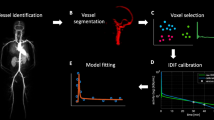
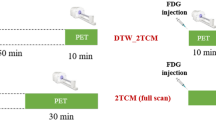
Abstract
Purpose
Positron emission tomography (PET) analysis of clinical studies is mostly restricted to qualitative evaluation. Quantitative analysis of PET studies is highly desirable to be able to compute an objective measurement of the process of interest in order to evaluate treatment response and/or compare patient data. But implementation of quantitative analysis generally requires the determination of the input function: the arterial blood or plasma activity which indicates how much tracer is available for uptake in the brain. The purpose of our work was to share with the community an open software tool that can assist in the estimation of this input function, and the derivation of a quantitative map from the dynamic PET study.
Methods
Arterial blood sampling during the PET study is the gold standard method to get the input function, but is uncomfortable and risky for the patient so it is rarely used in routine studies. To overcome the lack of a direct input function, different alternatives have been devised and are available in the literature. These alternatives derive the input function from the PET image itself (image-derived input function) or from data gathered from previous similar studies (population-based input function). In this article, we present ongoing work that includes the development of a software tool that integrates several methods with novel strategies for the segmentation of blood pools and parameter estimation.
Results
The tool is available as an extension to the 3D Slicer software. Tests on phantoms were conducted in order to validate the implemented methods. We evaluated the segmentation algorithms over a range of acquisition conditions and vasculature size. Input function estimation algorithms were evaluated against ground truth of the phantoms, as well as on their impact over the final quantification map. End-to-end use of the tool yields quantification maps with \({<}5\,\%\) relative error in the estimated influx versus ground truth on phantoms.
Conclusions
The main contribution of this article is the development of an open-source, free to use tool that encapsulates several well-known methods for the estimation of the input function and the quantification of dynamic PET FDG studies. Some alternative strategies are also proposed and implemented in the tool for the segmentation of blood pools and parameter estimation. The tool was tested on phantoms with encouraging results that suggest that even bloodless estimators could provide a viable alternative to blood sampling for quantification using graphical analysis. The open tool is a promising opportunity for collaboration among investigators and further validation on real studies.









Similar content being viewed by others
References
Cherry SR, Sorenson JA, Phelps ME (2012) Physics in nuclear medicine. Elsevier, Philadelphia
Zanotti-Fregonara P, Chen K, Liow JS, Fujita M, Innis RB (2011) Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab 31(10):1986–1998
Chen K, Bandy D, Reiman E, Huang SC, Lawson M, Feng D, Yun LS, Palant A (1998) Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. J Cereb Blood Flow Metab 18:716–723
Su KH, Wu LC, Liu RS, Wang SJ, Chen JC (2005) Quantification method in [18F]fluorodeoxyglucose brain positron emission tomography using independent component analysis. Nucl Med Commun 26:995–1004
Parker BJ, Feng D (2005) Graph-based Mumford–Shah segmentation of dynamic PET with application to input function estimation. IEEE Trans Nucl Sci 52:79–89
Zanotti-Fregonara Maroy R, Sureau F, Comtat C, Jan S, Syrota A, Tr’bossen R (2007) Noninvasive quantification of the cerebral metabolic rate of (18)F-FDG in dynamic brain PET studies using an image-derived input function. Eur J Nucl Med Mol Imaging 34(Suppl 2):S113
Zanotti-Fregonara P, Maroy R, Peyronneau MA, Trebossen R, Bottlaender M (2012) Minimally invasive input function for 2-\(^18\)F-fluoro-A-85380 brain PET studies. Eur J Nucl Med Mol Imaging 39(4):651–659
Soret M, Bacharach SL, Buvat I (2007) Partial-volume effect in PET tumor imaging. J Nucl Med 48(6):932–945
Naganawa M, Kimura Y, Ishii K, Oda K, Ishiwata K, Matani A (2005) Extraction of a plasma time-activity curve from dynamic brain PET images based on independent component analysis. IEEE Trans Biomed Eng 52(2):201–210
Hunter GJ, Hamberg LM, Alpert NM, Choi NC, Fischman AJ (1996) Simplied measurement of deoxyglucose utilization rate. J Nucl Med Soc publ Soc Nucl Med 37(6):950–955
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Zanotti-Fregonara P, Maroy R, Comtat C, Jan S, Gaura V, Bar-Hen A, Trebossen R (2009) Comparison of 3 methods of automated internal carotid segmentation in human brain PET studies: application to the estimation of arterial input function. J Nucl Med 50(3):461–467
Huang SC, Phelps ME, Homan EJ, Sideris K, Selin CJ, Kuhl DE (1980) Noninvasive determination of local cerebral metabolic rate of glucose in man. Am J Physiol 238(1):E69–E82
ASIM (2014) Analytic PET simulator Home Page (Online). University of Washington. http://depts.washington.edu/asimuw/
Thielemans Kris, Tsoumpas Charalampos, Mustafovic Sanida, Beisel Tobias, Aguiar Pablo, Dikaios Nikolaos, Jacobson Matthew W (2012) STIR: software for tomographic image reconstruction release 2. Phys Med Biol 57(4):867–883
Acknowledgments
This work was supported by Comision Sectorial de Investigacion Cientıfica (CSIC, Universidad de la Republica, Uruguay) under program “Proyecto de Inclusión Social.” The authors are grateful for the valuable comments of Dr. Rodolfo Ferrando, Dr. Andrés Damián, Dr. Patrick Dupont, and personal from CUDIM and Centro de Medicina Nuclear of Hospital de Clínicas.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Martín Bertran, Natalia Martínez, Guillermo Carbajal, Alicia Fernández and Alvaro Gómez declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Bertrán, M., Martínez, N., Carbajal, G. et al. An open tool for input function estimation and quantification of dynamic PET FDG brain scans. Int J CARS 11, 1419–1430 (2016). https://doi.org/10.1007/s11548-015-1307-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-015-1307-x