Abstract
Purpose
The main purpose of this work is to develop, apply, and evaluate an efficient approach for breast density estimation in magnetic resonance imaging data, which contain strong artifacts including intensity inhomogeneities.
Methods
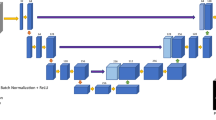
We present a pipeline for breast density estimation, which consists of intensity inhomogeneity correction, breast volume segmentation, nipple extraction, and fibroglandular tissue segmentation. For the segmentation steps, a well-known deep learning architecture is employed.
Results
The average Dice coefficient for the breast parenchyma is \(92.5\%\pm 0.011\), which outperforms the classical state-of-the-art approach by a margin of \(9\%\).
Conclusion
The proposed solution is accurate and highly efficient and has potential to be applied for big epidemiological data with thousands of participants.





Similar content being viewed by others
References
Altman DG, Bland JM (1994) Diagnostic tests. 1: sensitivity and specificity. Br Med J: BMJ 308(6943):1552
Bland JM, Altman D (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327(8476):307–310
Dalmış MU, Litjens G, Holland K, Setio A, Mann R, Karssemeijer N, Gubern-Mérida A (2017) Using deep learning to segment breast and fibroglandular tissue in mri volumes. Med Phys 44(2):533–546
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Eberl MM, Fox CH, Edge SB, Carter CA, Mahoney MC (2006) Bi-rads classification for management of abnormal mammograms. J Am Board Fam Med 19(2):161–164
Gonzalez RC, Woods RE (2002) Digital image processing. Prentice Hall, Upper Saddle River
Greenspan H, van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35(5):1153–1159
Gubern-Merida A, Kallenberg M, Mann RM, Marti R, Karssemeijer N (2015) Breast segmentation and density estimation in breast MRI: a fully automatic framework. IEEE J Biomed Health Inform 19(1):349–357
Hou Z (2006) A review on MR image intensity inhomogeneity correction. Int J Biomed Imaging 2006:1e11
Ivanovska T, Gloger LWO, Hahn HK, Völzke H, Hegenscheid K (2015) Sliding level set-based boundary: fully automated dense breast segmentation in native MR mammograms. In: Proceedings of MICCAI-BIA p 57
Ivanovska T, Laqua R, Wang L, Liebscher V, Völzke H, Hegenscheid K (2014) A level set based framework for quantitative evaluation of breast tissue density from mri data. PLoS ONE 9(11):1–19
Ivanovska T, Laqua R, Wang L, Schenk A, Yoon JH, Hegenscheid K, Völzke H, Liebscher V (2016) An efficient level set method for simultaneous intensity inhomogeneity correction and segmentation of mr images. Comput Med Imaging Graph 48:9–20
Jentschke TG, Hegenscheid K, Völzke H, Wörgötter F, Ivanovska T (2018) Segmentierung von brustvolumina in magnetresonanztomographiedaten unter der verwendung von deep learning. In: Proceedings of workshop Bildverarbeitung in der Medizin (BVM)
John U, Hensel E, Lüdemann J, Piek M, Sauer S, Adam C, Born G, Alte D, Greiser E, Haertel U, Hense HW, Haerting J, Willich S, Kessler C (2001) Study of health in pomerania (ship): a health examination survey in an east german region: objectives and design. Sozial-und Präventivmedizin 46(3):186–194
McCormack VA, dos Santos Silva I (2006) Breast density and parenchymal patterns as markers of breast cancer risk: a meta-analysis. Cancer Epidemiol Prev Biomark 15(6):1159–1169
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, pp 565–571
Razavi M, Wang L, Gubern-Mérida A, Ivanovska T, Laue H, Karssemeijer N, Hahn HK (2015) Towards accurate segmentation of fibroglandular tissue in breast MRI using fuzzy c-means and skin-folds removal. In: International conference on image analysis and processing. Springer, pp 528–536
Rosset A, Spadola L, Ratib O (2004) Osirix: an open-source software for navigating in multidimensional dicom images. J Digit Imaging 17(3):205–216
Schmidhuber J (2015) Deep learning in neural networks: an overview. Neural Netw 61:85–117
Smith TB, Nayak KS (2010) Mri artifacts and correction strategies. Imaging Med 2(4):445–457
Tice JA, Cummings SR, Ziv E, Kerlikowske K (2005) Mammographic breast density and the gail model for breast cancer risk prediction in a screening population. Breast Cancer Res Treat 94(2):115–122
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4itk: improved n3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Vachon CM, Kuni CC, Anderson K, Anderson VE, Sellers TA (2000) Association of mammographically defined percent breast density with epidemiologic risk factors for breast cancer (united states). Cancer Causes Control 11(7):653–662
Vincent R (2006) Brainweb: simulated brain database. Available online at http://www.bic.mni.mcgill.ca/brainweb/
Vovk U, Pernus F, Likar B (2007) A review of methods for correction of intensity inhomogeneity in mri. IEEE Trans Med Imaging 26(3):405–421
Zhuo J, Gullapalli RP (2006) Mr artifacts, safety, and quality control. Radiographics 26(1):275–297
Acknowledgements
This study is developed within the project, which is partially supported by the German Research Foundation, Project No. IV 161/4-1, DA 1810/2-1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Informed consent was obtained from all individuals included in the study. This study was a subproject of the population-based Study of Health in Pomerania (SHIP). SHIP was conducted in the Northeast German federal state of Mecklenburg-Western Pomerania. Written informed consent was obtained separately for study inclusion and MR imaging.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards. The SHIP was conducted as approved by the local Institutional Review Board at Greifswald University Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ivanovska, T., Jentschke, T.G., Daboul, A. et al. A deep learning framework for efficient analysis of breast volume and fibroglandular tissue using MR data with strong artifacts. Int J CARS 14, 1627–1633 (2019). https://doi.org/10.1007/s11548-019-01928-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-01928-y