Abstract
Purpose
The World Health Organization (WHO) grading system of pancreatic neuroendocrine tumor (PNET) plays an important role in the clinical decision. The rarity of PNET often negatively affects the radiological application of deep learning algorithms due to the low availability of radiological images. We tried to investigate the feasibility of predicting WHO grades of PNET on contrast-enhanced magnetic resonance (MR) images by deep learning algorithms.
Materials and methods
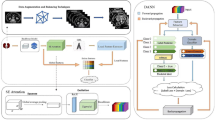
Ninety-six patients with PNET underwent preoperative contrast-enhanced MR imaging. Fivefold cross-validation was used in which five iterations of training and validation were performed. Within every iteration, on the training set augmented by synthetic images generated from generative adversarial network (GAN), a convolutional neural network (CNN) was trained and its performance was evaluated on the paired internal validation set. Finally, the trained CNNs from cross-validation and their averaged counterpart were separately assessed on another ten patients from a different external validation set.
Results
Averaging the results across the five iterations in the cross-validation, for the CNN model, the average accuracy was 85.13% ± 0.44% and micro-average AUC was 0.9117 ± 0.0053. Evaluated on the external validation set, the average accuracy of the five trained CNNs ranges between 79.08 and 82.35%, and the range of micro-average AUC was between 0.8825 and 0.8932. The average accuracy and micro-average AUC of the averaged CNN were 81.05% and 0.8847, respectively.
Conclusion
Synthetic images generated from GAN could be used to alleviate the difficulty of radiological image collection for uncommon disease like PNET. With the help of GAN, the CNN showed the potential to predict the WHO grades of PNET on contrast-enhanced MR images.





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Halfdanarson TR, Rabe KG, Rubin J, Petersen GM (2008) Pancreatic neuroendocrine tumors (PNETs): incidence, prognosis and recent trend toward improved survival. Ann Oncol 19:1727–1733. https://doi.org/10.1093/annonc/mdn351
Ramage JK, Ahmed A, Ardill J, Bax N, Breen DJ, Caplin ME, Corrie P, Davar J, Davies AH, Lewington V, Meyer T, Newell-Price J, Poston G, Reed N, Rockall A, Steward W, Thakker RV, Toubanakis C, Valle J, Verbeke C, Grossman AB, UK and Ireland Neuroendocrine Tumour Society (2011) Guidelines for the management of gastroenteropancreatic neuroendocrine (including carcinoid) tumours (NETs). Gut 61:6–32. https://doi.org/10.1136/gutjnl-2011-300831
Klimstra DS, Modlin IR, Coppola D, Lloyd RV, Suster S (2010) The pathologic classification of neuroendocrine tumors: a review of nomenclature, grading, and staging systems. Pancreas 39:707–712. https://doi.org/10.1097/MPA.0b013e3181ec124e
Bettini R, Partelli S, Boninsegna L, Capelli P, Crippa S, Pederzoli P, Scarpa A, Falconi M (2011) Tumor size correlates with malignancy in nonfunctioning pancreatic endocrine tumor. Surgery 150:75–82. https://doi.org/10.1016/j.surg.2011.02.022
Hwang EJ, Lee JM, Yoon JH, Kim JH, Han JK, Choi BI, Lee KB, Jang JY, Kim SW, Nickel MD, Kiefer B (2014) Intravoxel incoherent motion diffusion-weighted imaging of pancreatic neuroendocrine tumors: prediction of the histologic grade using pure diffusion coefficient and tumor size. Invest Radiol 49:396–402. https://doi.org/10.1097/RLI.0000000000000028
Pereira JAS, Rosado E, Bali M, Metens T, Chao S-L (2015) Pancreatic neuroendocrine tumors: correlation between histogram analysis of apparent diffusion coefficient maps and tumor grade. Abdom Imaging 40:3122–3128. https://doi.org/10.1007/s00261-015-0524-7
Cappelli C, Boggi U, Mazzeo S, Cervelli R, Campani D, Funel N, Contillo BP, Bartolozzi C (2015) Contrast enhancement pattern on multidetector CT predicts malignancy in pancreatic endocrine tumours. Eur Radiol 25:751–759. https://doi.org/10.1007/s00330-014-3485-2
De Robertis R, Cingarlini S, Tinazzi Martini P, Ortolani S, Butturini G, Landoni L, Regi P, Girelli R, Capelli P, Gobbo S, Tortora G, Scarpa A, Pederzoli P, D’Onofrio M (2017) Pancreatic neuroendocrine neoplasms: magnetic resonance imaging features according to grade and stage. World J Gastroenterol 23:275–285. https://doi.org/10.3748/wjg.v23.i2.275
Lotfalizadeh E, Ronot M, Wagner M, Cros J, Couvelard A, Vullierme MP, Allaham W, Hentic O, Ruzniewski P, Vilgrain V (2017) Prediction of pancreatic neuroendocrine tumour grade with MR imaging features: added value of diffusion-weighted imaging. Eur Radiol 27:1748–1759. https://doi.org/10.1007/s00330-016-4539-4
Kulali F, Semiz-Oysu A, Demir M, Segmen-Yilmaz M, Bukte Y (2018) Role of diffusion-weighted MR imaging in predicting the grade of nonfunctional pancreatic neuroendocrine tumors. Diagn Interv Imaging 99:301–309. https://doi.org/10.1016/j.diii.2017.10.012
De Robertis R, Maris B, Cardobi N, Tinazzi Martini P, Gobbo S, Capelli P, Ortolani S, Cingarlini S, Paiella S, Landoni L, Butturini G, Regi P, Scarpa A, Tortora G, D’Onofrio M (2018) Can histogram analysis of MR images predict aggressiveness in pancreatic neuroendocrine tumors? Eur Radiol 28:2582–2591. https://doi.org/10.1007/s00330-017-5236-7
Cicero M, Bilbily A, Colak E, Dowdell T, Gray B, Perampaladas K, Barfett J (2017) Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol 52:281–287. https://doi.org/10.1097/RLI.0000000000000341
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582. https://doi.org/10.1148/radiol.2017162326
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896. https://doi.org/10.1148/radiol.2017170706
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88. https://doi.org/10.1016/j.media.2017.07.005
Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A (2017) Deep learning: a primer for radiologists. Radiographics 37:2113–2131. https://doi.org/10.1148/rg.2017170077
Creswell A, White T, Dumoulin V, Arulkumaran K, Sengupta B, Bharath AA (2018) Generative adversarial networks: an overview. IEEE Signal Process Mag 35:53–65. https://doi.org/10.1109/MSP.2017.2765202
Jeon SK, Lee JM, Joo I, Lee ES, Park HJ, Jang JY, Ryu JK, Lee KB, Han JK (2017) Nonhypervascular pancreatic neuroendocrine tumors: differential diagnosis from pancreatic ductal adenocarcinomas at MR imaging-retrospective cross-sectional study. Radiology 284:77–87. https://doi.org/10.1148/radiol.2016160586
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671. https://doi.org/10.1038/nmeth.2089
Radford A, Metz L, Chintala S (2015) Unsupervised representation learning with deep convolutional generative adversarial networks. http://arxiv.org/pdf/1511.06434v2. Accessed 30 April 2019
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45:427–437. https://doi.org/10.1016/j.ipm.2009.03.002
Wei R, Wang J, Jia W (2019) MultiROC: calculating and visualizing ROC and PR curves across multi-class classifications. https://cran.r-project.org/web/packages/multiROC/index.html. Accessed 30 April 2019
Pedregosa F, Varoquaux G, Gramfort A, Michel V (2019) Receiver operating characteristic (ROC). https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.html. Accessed 30 April 2019
Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Man´e D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Vi´egas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, Zheng X (2016) TensorFlow: large-scale machine learning on heterogeneous distributed systems. http://arxiv.org/pdf/1603.04467v2. Accessed 30 April 2019
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. http://arxiv.org/pdf/1412.6980v9. Accessed 30 April 2019
Buda M, Maki A, Mazurowski MA (2018) A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw 106:249–259. https://doi.org/10.1016/j.neunet.2018.07.011
Haixiang G, Yijing L, Shang J, Mingyun G, Yuanyue H, Bing G (2017) Learning from class-imbalanced data: review of methods and applications. Expert Syst Appl 73:220–239. https://doi.org/10.1016/j.eswa.2016.12.035
Taylor L, Nitschke G (2017) Improving deep learning using generic data augmentation. http://arxiv.org/pdf/1708.06020v1. Accessed 30 April 2019
Xue D-X, Zhang R, Feng H, Wang Y-L (2016) CNN-SVM for microvascular morphological type recognition with data augmentation. J Med Biol Eng 36:755–764. https://doi.org/10.1007/s40846-016-0182-4
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial networks. http://arxiv.org/pdf/1406.2661v1. Accessed 30 April 2019
Acknowledgements
We thank Dr. Feixiang Hu and Prof. Weijun Peng from Fudan University Shanghai Cancer Center, Yang Zhou from Fudan University Zhongshan Hospital, for their kind help in this study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Research involving human participants and/or animals
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
For this type of study, formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
In this article, Fudan University Zhongshan Hospital was referred to as HOSPITAL ONE and Fudan University Shanghai Cancer Center was referred to as HOSPITAL TWO.
Rights and permissions
About this article
Cite this article
Gao, X., Wang, X. Deep learning for World Health Organization grades of pancreatic neuroendocrine tumors on contrast-enhanced magnetic resonance images: a preliminary study. Int J CARS 14, 1981–1991 (2019). https://doi.org/10.1007/s11548-019-02070-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-02070-5