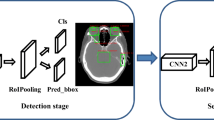
Abstract
Purpose
Nasopharyngeal carcinoma (NPC) is a category of tumors with high incidence in head-and-neck (H&N) body region, and the diagnosis and treatment planning are usually conducted by radiologists manually, which is tedious, time-consuming and unrepeatable. In this paper, we integrated different stages of this process and proposed a computer-aided framework to realize automatic detection, tumor region and sub-region segmentation, and visualization of NPC, which are usually investigated separately in literatures.
Methods
Multi-modality images are utilized in the framework. Firstly, NPC is detected by a convolutional neural network (CNN) on computed tomography (CT) scans. Then, NPC area is segmented from magnetic resonance imaging (MRI) images by using a multi-modality MRI fusion network. Thirdly, NPC sub-regions with different metabolic activities are divided on CT images of the same patient via an adaptive threshold algorithm. Finally, 3D surface model of NPC is generated for observing its shape, size, and location in the head region. The proposed method is compared with other algorithms by evaluation on the volumes and shapes of detected NPCs.
Results
Experiments are conducted on CT images of 130 NPC patients and 102 subjects without NPC and MRI images of 149 NPC patients, among which 52 subjects are overlapped with both CT and MRI images. The reference for evaluation is generated by three experienced radiologists. The results demonstrated that our utilized models outperform other strategies with detection accuracy 0.882 and Dice similarity coefficient 0.719 for NPC segmentation. Sub-region division and the 3D visualized models show great acceptability in clinical usage.
Conclusion
The remarkable performance indicated the potential of our framework in alleviating workload of radiologist. Furthermore, the combined usage of multi-modality images is able to generate reliable segmentations of NPC area and sub-regions, which provide evidence to judge the heterogeneity among patients and guide the dose painting for radiation therapy.









Similar content being viewed by others
References
Mohammed MA, Ghani MKA, Hamed RI, Ibrahim DA (2017) Review on nasopharyngeal carcinoma: concepts, methods of analysis, segmentation, classification, prediction and impact: a review of the research literature. J Comput Sci 21:283–298. https://doi.org/10.1016/j.jocs.2017.03.021
Chang ET, Adami HO (2006) The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 15(10):1765–1777. https://doi.org/10.1158/1055-9965.EPI-06-0353
Chong VF, Fan YF, Khoo JB (1996) Nasopharyngeal carcinoma with intracranial spread: CT and MR characteristics. J Comput Assist Tomogr 20(4):563–569. https://doi.org/10.1097/00004728-199607000-00012
Rasch C, Keus R, Pameijer FA, Koops W, de Ru V, Muller S, Touw A, Bartelink H, van Herk M, Lebesque JV (1997) The potential impact of CT-MRI matching on tumor volume delineation in advanced head and neck cancer. Int J Radiat Oncol Biol Phys 39(4):841–848. https://doi.org/10.1016/s0360-3016(97)00465-3
Razek AAKA, King A (2012) MRI and CT of nasopharyngeal carcinoma. Am J Roentgenol 198(1):11–18. https://doi.org/10.2214/AJR.11.6954
Razek AAKA, Kamal E (2013) Nasopharyngeal carcinoma: correlation of apparent diffusion coefficient value with prognostic parameters. Radiol Med 118:534–539. https://doi.org/10.1007/s11547-012-0890-x
Tatanun C, Ritthipravat P, Bhongmakapat T, Tuntiyatorn L (2010) Automatic segmentation of nasopharyngeal carcinoma from CT images: Region growing based technique. In: 2010 2nd International conference on signal processing systems, vol 2. IEEE, pp 537–541. https://doi.org/10.1109/ICSPS.2010.5555663
Chanapai W, Bhongmakapat T, Tuntiyatorn L, Ritthipravat P (2012) Nasopharyngeal carcinoma segmentation using a region growing technique. Int J Comput Assist Radiol Surg 7(3):413–422. https://doi.org/10.1007/s11548-011-0629-6
Lee FK, Yeung DK, King AD, Leung S, Ahuja A (2005) Segmentation of nasopharyngeal carcinoma (NPC) lesions in MR images. Intl J Radiat Oncol* Biol* Phys 61(2):608–620. https://doi.org/10.1016/j.ijrobp.2004.09.024
Chanapai W, Ritthipravat P (2009) Adaptive thresholding based on SOM technique for semi-automatic NPC image segmentation. In 2009 International conference on machine learning and applications. IEEE, pp 504–508. https://doi.org/10.1109/ICMLA.2009.135
Huang KW, Zhao ZY, Gong Q, Zha J, Chen L, Yang R (2015) Nasopharyngeal carcinoma segmentation via HMRF-EM with maximum entropy. In: 2015 37th Annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE, 2968–2972. https://doi.org/10.1109/EMBC.2015.7319015
Fitton I, Cornelissen S, Duppen JC, Steenbakkers R, Peeters S, Hoebers F, Kaanders JH, Nowak P, Rasch CR, van Herk M (2011) Semi-automatic delineation using weighted CT-MRI registered images for radiotherapy of nasopharyngeal cancer. Med Phys 38(8):4662–4666. https://doi.org/10.1118/1.3611045
Zhou J, Chan KL, Xu P, Chong VF (2006) Nasopharyngeal carcinoma lesion segmentation from MR images by support vector machine. In: 3rd IEEE international symposium on biomedical imaging: nano to macro. IEEE, pp 1364–1367. https://doi.org/10.1109/ISBI.2006.1625180
Zhang J, Ma KK, Er MH, Chong V (2004) Tumor segmentation from magnetic resonance imaging by learning via one-class support vector machine. In: International workshop on advanced image technology (IWAIT’04), pp 207–211
Zhou J, Chong V, Lim TK, Houng J (2002) MRI tumor segmentation for nasopharyngeal carcinoma using knowledge-based fuzzy clustering. Int J Inf Technol 8(2):36–45
Feng A, Chen Z, Wu X, Ma Z (2017) From convolutional to recurrent: Case study in nasopharyngeal carcinoma segmentation. In: 2017 International conference on the frontiers and advances in data science (FADS). IEEE, pp 18–22. https://doi.org/10.1109/FADS.2017.8253187
Ma Z, Wu X, Zhou J (2017) Automatic nasopharyngeal carcinoma segmentation in MR images with convolutional neural networks. In: 2017 International conference on the frontiers and advances in data science (FADS). IEEE, pp 147–150. https://doi.org/10.1109/FADS.2017.8253215
Ma Z, Wu X, Song Q, Luo Y, Wang Y, Zhou J (2018) Automated nasopharyngeal carcinoma segmentation in magnetic resonance images by combination of convolutional neural networks and graph cut. Exp Ther Med 16(3):2511–2521. https://doi.org/10.3892/etm.2018.6478
Ma Z, Wu X, Sun S, Xia C, Yang Z, Li S, Zhou J (2018) A discriminative learning based approach for automated nasopharyngeal carcinoma segmentation leveraging multi-modality similarity metric learning. In: 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018). IEEE, pp 813–816. https://doi.org/10.1109/ISBI.2018.8363696
Tseng KL, Lin YL, Hsu W, Huang CY (2017) Joint sequence learning and cross-modality convolution for 3D biomedical segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 6393–6400. https://doi.org/10.1109/CVPR.2017.398
Valindria VV, Pawlowski N, Rajchl M, Lavdas I, Aboagye EO, Rockall AG, Rueckert D, Glocker B (2018) Multi-modal learning from unpaired images: Application to multi-organ segmentation in CT and MRI. In: Proceedings of the IEEE winter conference on applications of computer vision (WACV), pp 547–556. https://doi.org/10.1109/WACV.2018.00066
Dolz J, Gopinath K, Yuan J, Lombaert H, Desrosiers C, Ayed IB (2018) Hyperdense-net: a hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans Med Imaging 38(5):1116–1126. https://doi.org/10.1109/TMI.2018.2878669
Dolz J, Desrosiers C, Ayed IB (2018) Ivd-net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNET. In: Proceedings of the international workshop and challenge on computational methods and clinical applications for spine imaging, pp 130–143. https://doi.org/10.1007/978-3-030-13736-6_11
Tang P, Zu C, Hong M, Yan R, Peng X, Xiao J, Wu X, Zhou J, Zhou L, Wang Y (2021) DA-DSUnet: dual attention-based dense SU-net for automatic head-and-neck tumor segmentation in MRI images. Neurocomputing 435(7):103–113. https://doi.org/10.1016/j.neucom.2020.12.085
Men K, Chen X, Yang B, Zhu J, Yi J, Wang S, Li Y, Dai J (2021) Automatic segmentation of three clinical target volumes in radiotherapy using lifelong learning. Radiother Oncol 157:1–7. https://doi.org/10.1016/j.radonc.2020.12.034
Guo F, Shi C, Li X, Wu X, Zhou J, Lv J (2020) Image segmentation of nasopharyngeal carcinoma using 3D CNN with long-range skip connection and multi-scale feature pyramid. Soft Comput 24(16):12671–12680. https://doi.org/10.1007/s00500-020-04708-y
Serganova I, Doubrovin M, Vider J, Ponomarev V, Soghomonyan S, Beresten T, Ageyeva L, Serganov A, Cai S, Balatoni J, Blasberg R, Gelovani J (2004) Molecular imaging of temporal dynamics and spatial heterogeneity of hypoxia-inducible factor-1 signal transduction activity in tumors in living mice. Cancer Res 64:6101–6108. https://doi.org/10.1158/0008-5472.CAN-04-0842
Baumann M, Krause M, Overgaard J, Debus J, Bentzen SM, Daartz J, Richter C, Zips D, Bortfeld T (2016) Radiation oncology in the era of precision medicine. Nat Rev Cancer 16:234–249. https://doi.org/10.1038/nrc.2016.18
Wu J, Gong G, Cui Y, Li R (2016) Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging 44(5):1107–1115. https://doi.org/10.1002/jmri.25279
Hanahan D, Folkman J (1996) Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 86(3):353–364. https://doi.org/10.1016/S0092-8674(00)80108-7
Mang A, Bakas S, Subramanian S, Davatzikos C, Biros G (2020) Integrated biophysical modeling and image analysis: application to neuro-oncology. Annu Rev Biomed Eng 22:309–341. https://doi.org/10.1146/annurev-bioeng-062117-121105
Devic S (2013) Towards biological target volumes definition for radiotherapy treatment planning: Quo Vadis PET/CT? J Nucl Med Radiat Ther 4(3):1–10. https://doi.org/10.4172/2155-9619.1000158
Farhidzadeh H, Kim JY, Scott JG, Goldgof DB, Hall LO, Harrison LB (2016) Classification of progression free survival with nasopharyngeal carcinoma tumors. In: Medical imaging 2016: computer-aided diagnosis, international society for optics and photonics, vol 9785, p 97851I. https://doi.org/10.1117/12.2216976
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66. https://doi.org/10.1109/TSMC.1979.4310076
Ong CK, Chong VFH (2010) Imaging in the diagnosis and staging of carcinoma of nasopharynx nasopharyngeal cancer. Springer, Berlin
Wei L, GuangFeng D, RiJie T (2012) Relationship between CT enhancement and T staging of nasopharyngeal carcinoma. Guangdong Med J 33(6):773–775
Glastonbury C (2007) Nasopharyngeal carcinoma: the role of magnetic resonance imaging in diagnosis, staging, treatment, and follow-up. Top Magn Reson Imaging 18(4):225–235. https://doi.org/10.1097/RMR.0b013e3181572b3a
Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence, vol 4278–4284
Chen H, Qi Y, Yin Y, Li T, Gong G, Wang L (2020) MMFNet: A multimodality MRI fusion network for segmentation of nasopharyngeal carcinoma. Neurocomputing 394(21):27–40. https://doi.org/10.1016/j.neucom.2020.02.002
Huang YJ, Dou Q, Wang ZX, Liu LZ, Jin Y, Li CF, Wang L, Chen H, Xu RH (2020) 3d roi-aware u-net for accurate and efficient colorectal tumor segmentation. IEEE Trans Cybern. https://doi.org/10.1109/TCYB.2020.2980145
Leger S, Zwanenburg A, Leger K, Lohaus F, Linge A, Schreiber A, Kalinauskaite G, Tinhofer I, Guberina N, Guberina M, Balermpas P, von der Grün J, Ganswindt U, Belka C, Peeken JC, Combs SE, Boeke S, Zips D, Richter C, Krause M, Baumann M, Troost EGC, Löck S (2020) Comprehensive analysis of tumour sub-volumes for radiomic risk modelling in locally advanced HNSCC. Cancers 12(10):3047. https://doi.org/10.3390/cancers12103047
Wolf I, Vetter M, Wegner I, Böttger T, Nolden M, Schöbinger M, Hastenteufel M, Kunert T, Meinzer HP (2005) The medical imaging interaction toolkit. Med Image Anal 9(6):594–604. https://doi.org/10.1016/j.media.2005.04.005
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: 2015 IEEE conference on computer vision and pattern recognition (CVPR), pp 1–9. https://doi.org/10.1109/CVPR.2015.7298594
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3D U-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computerassisted intervention, pp 424–432. https://doi.org/10.1007/978-3-319-46723-8_49
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78. https://doi.org/10.1016/j.media.2016.10.004
Funding
This work was funded in part by grants from the National Key R&D Program of China (Grant No. 2017YFB1302900), Natural Science Foundation of Shandong Province (ZR2020MH227), and SJTU Translational Medicine Cross Research Fund (YG2019ZDA26).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This retrospective study was conducted following ethical approval from the Institutional Review Board at the Hospital of the Shandong Cancer Hospital and Institute. This article does not contain any studies with animals performed by any of the authors. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qi, Y., Li, J., Chen, H. et al. Computer-aided diagnosis and regional segmentation of nasopharyngeal carcinoma based on multi-modality medical images. Int J CARS 16, 871–882 (2021). https://doi.org/10.1007/s11548-021-02351-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02351-y