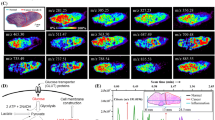
Abstract
Purpose
Intraoperative assessment of surgical margins is important for reducing the rate of revisions in breast conserving surgery for palpable malignant tumors. The hypothesis was that metabolomics methods, based on mass spectrometry, could find patterns of relative abundances of molecules that distinguish clusters of benign tissue and cancer in surgical resections.
Methods
Excisions from 8 patients were used to acquire 112,317 mass spectrometry signals by desorption electrospray ionization. A process of nonnegative matrix factorization and graph decomposition produced clusters that were approximated as affine spaces. Each signal’s distance to the affine space of a cluster was used to visualize the clustering.
Results
The distance maps were superior to binary clustering in identifying cancer regions. They were particularly effective at finding cancer regions that were discontinuously distributed within benign tissue.
Conclusions
Desorption electrospray ionization mass spectrometry, which has been shown to be useful intraoperatively, can acquire signals that distinguish malignant from benign breast tissue in surgically excised tumors. The method may be suitable for real-time surgical decisions based on cancer margins.








Similar content being viewed by others
References
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, Jemal A (2019) Breast cancer statistics. CA Cancer J Clin 69(6):438–451
Havel L, Naik H, Ramirez L, Morrow M, Landercasper J (2019) Impact of the SSO-ASTRO margin guideline on rates of re-excision after lumpectomy for breast cancer: a meta-analysis. Ann Surg Oncol 26(5):1238–1244
van Dooijeweert C, van Diest PJ, Willems SM, Kuijpers CCHJ, van der Wall E, Overbeek LIH, Deckers IAG (2020) Significant inter- and intra-laboratory variation in grading of invasive breast cancer: a nationwide study of 33,043 patients in the Netherlands. Int J Cancer 146(3):769–780
McCartney A, Vignoli A, Biganzoli L, Love R, Tenori L, Luchinat C, Di Leo A (2018) Metabolomics in breast cancer: a decade in review. Cancer Treat Rev 67:88–96
van Hove ERA, Smith DF, Heeren RMA (2010) A concise review of mass spectrometry imaging. J Chromatogr A 1217(25):3946–3954
Takats Z, Wiseman JM, Gologan B, Cooks RG (2004) Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306(5695):471–473
Pirro V, Alfaro CM, Jarmusch AK, Hattab EM, Cohen-Gadol AA, Cooks RG (2017) Intraoperative assessment of tumor margins during glioma resection by desorption electrospray ionization-mass spectrometry. Proc Natl Acad Sci 114(26):6700–6705
Guenther S, Muirhead LJ, Speller AV, Golf O, Strittmatter N, Ramakrishnan R, Goldin RD, Jones E, Veselkov K, Nicholson J, Darzi A, Takats Z (2015) Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res 75(9):1828–1837
Race AM, Bunch J (2015) Optimisation of colour schemes to accurately display mass spectrometry imaging data based on human colour perception. Anal Bioanal Chem 407(8):2047–2054
Jolliffe IT, Cadima J (2016) Principal component analysis: a review and recent developments. Phil Trans R Soc A 374(2065):20150202
Vargo-Gogola T, Rosen JM (2007) Modelling breast cancer: one size does not fit all. Nat Rev Cancer 7(9):659–672
Santoro AL, Drummond RD, Silva IT, Ferreira SS, Juliano L, Vendramini PH, da Costa Lemos MB, Eberlin MN, Andrade VP (2020) In situ DESI-MSI lipidomic profiles of breast cancer molecular subtypes and precursor lesions. Cancer Res 80(6):1246–1257
Jeffries N (2005) Algorithms for alignment of mass spectrometry proteomic data. Bioinformatics 21(14):3066–3073
Lee DD, Seung HS (1999) Learning the parts of objects by non-negative matrix factorization. Nature 401(6755):788–791
Paatero P, Tapper U (1994) Positive matrix factorization: a non-negative factor model with optimal utilization of error estimates of data values. Environmetrics 5(2):111–126
Kim J, He Y, Park H (2014) Algorithms for nonnegative matrix and tensor factorizations: a unified view based on block coordinate descent framework. J Global Optim 58(2):285–319
Boutsidis C, Gallopoulos E (2008) SVD based initialization: a head start for nonnegative matrix factorization. Pattern Recogn 41(4):1350–1362
Elhamifar E, Vidal R (2013) Sparse subspace clustering: algorithm, theory, and applications. IEEE Trans Pattern Anal Mach Intell 35(11):2765–2781
Pourkamali-Anaraki F, Folberth J, Becker S (2020) Efficient solvers for sparse subspace clustering. Signal Process 172:107548
Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A (2018) HMDB 4.0: the human metabolome database for 2018. Nucl Acids Res 46(D1):D608–D617
Ester M, Kriegel HP, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. Mach Learn Knowl Discov Databases 96:226–231
Pluim JP, Maintz JA, Viergever MA (2003) Mutual-information-based registration of medical images: a survey. IEEE Trans Med Imaging 22(8):986–1004
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Menendez JA, Lupu R (2007) Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7(10):763–777
Parry RM, Galhena AS, Gamage CM, Bennett RV, Wang MD, Fernández FM (2013) omnispect: an open MATLAB-based tool for visualization and analysis of matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry images. J Am Soc Mass Spectrom 24(4):646–649
Paine MR, Kim J, Bennett RV, Parry RM, Gaul DA, Wang MD, Matzuk MM, Fernández FM (2016) Whole reproductive system non-negative matrix factorization mass spectrometry imaging of an early-stage ovarian cancer mouse model. PLoS One 11(5):e0154837
Trindade GF, Abel ML, Watts JF (2017) Non-negative matrix factorisation of large mass spectrometry datasets. Chemometr Intell Lab Syst 163:76–85
Margulis K, Chiou AS, Aasi SZ, Tibshirani RJ, Tang JY, Zare RN (2018) Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc Natl Acad Sci 115(25):6347–6352
Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, Jarmusch AK, Ligon KL, Jolesz FA, Golby AJ, Santagatac S, Agar NYR, Cooks RG (2013) Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci 110(5):1611–1616
Acknowledgements
Nicole Morse contributed to the DESI-MS data acquisition when she was a graduate student. We are grateful to Emma Ritcey and Tyler Mainguy for their computational assistance in preprocessing the data. This work was supported in part by the Natural Sciences and Engineering Research Council of Canada under Grant RGPIN-2018-04430, and in part by a Vector Scholarship in Artificial Intelligence, provided through the Vector Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
This study was conducted in accordance with the principles outlined in the Declaration of Helsinki for the use of human tissue.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Theriault, R.L., Kaufmann, M., Ren, K.Y.M. et al. Metabolomics patterns of breast cancer tumors using mass spectrometry imaging. Int J CARS 16, 1089–1099 (2021). https://doi.org/10.1007/s11548-021-02387-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02387-0