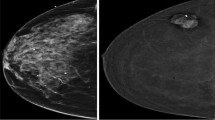
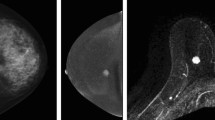
Abstract
Purpose
CESM (contrast-enhanced spectral mammography) is an efficient tool for detecting breast cancer because of its image characteristics. However, among most deep learning-based methods for breast cancer classification, few models can integrate both its multiview and multimodal features. To effectively utilize the image features of CESM and thus help physicians to improve the accuracy of diagnosis, we propose a multiview multimodal network (MVMM-Net).
Methods
The experiment is carried out to evaluate the in-house CESM images dataset taken from 95 patients aged 21–74 years with 760 images. The framework consists of three main stages: the input of the model, image feature extraction, and image classification. The first stage is to preprocess the CESM to utilize its multiview and multimodal features effectively. In the feature extraction stage, a deep learning-based network is used to extract CESM images features. The last stage is to integrate different features for classification using the MVMM-Net model.
Results
According to the experimental results, the proposed method based on the Res2Net50 framework achieves an accuracy of 96.591%, sensitivity of 96.396%, specificity of 96.350%, precision of 96.833%, F1_score of 0.966, and AUC of 0.966 on the test set. Comparative experiments illustrate that the classification performance of the model can be improved by using multiview multimodal features.
Conclusion
We proposed a deep learning classification model that combines multiple features of CESM. The results of the experiment indicate that our method is more precise than the state-of-the-art methods and produces accurate results for the classification of CESM images.







Similar content being viewed by others
References
Farzad R, Gordon F, Sahar T, Mahnaz N, Amir A, Soodabeh S (2020) Role of regulatory miRNAS of the pi3K/AKT signaling pathway in the pathogenesis of breast cancer. Gene 737(144459):02
Siegel R, Miller K, Fuchs H, Jemal A (2021) Cancer statistics. CA A Cancer J Clin 71(1):7–33
Hiba C, Hamid Z, Omar A (2020) Multi-label transfer learning for the early diagnosis of breast cancer. Neurocomputing 392:168–180
Martin D, Tobias DZ, Wolfram S, Birgit A, Florian K, Werner J, Clarisse D, Willi O, Michael H, Christian M (2015) Dual-energy contrast-enhanced spectral mammography (CESM). Arch Gynecol Obstet 292(4):739–747
Lisa H, Martin D, Christoph J, Elena E, Julia H (2019) Contrast-enhanced spectral mammography with a compact synchrotron source. PLoS ONE 14(10):
James JJ, Tennant SL (2018) Contrast-enhanced spectral mammography (CESM). Clin Radiol 73(8):715–723
Lobbes MBI, Smidt ML, Houwers J, Tjan-Heijnen VC, Wildberger JE (2013) Contrast enhanced mammography: techniques, current results, and potential indications. Clin Radiol 68(9):935–944
Cheung YC, Tsai HP, Lo YF, Ueng SH, Huang PC, Chen SC (2016) Clinical utility of dual-energy contrast-enhanced spectral mammography for breast microcalcifications without associated mass: a preliminary analysis. Eur Radiol 26(4):1082–1089
Patel BK, Ranjbar S, Wu T, Pockaj BA, Li J, Zhang N, Lobbes M, Zhang B, Mitchell JR (2018) Computer-aided diagnosis of contrast-enhanced spectral mammography: a feasibility study. Eur J Radiol 98(3):207–213
Gopichandh D, Bhavika P, Faranak A, Morteza H, Jing L, Teresa W, Bin Z (2018) Classification of breast masses using a computer-aided diagnosis scheme of contrast enhanced digital mammograms. Ann Biomed Eng 46(9):1419–1431
Dromain C, Thibault F, Diekmann F, Fallenberg EM, Jong RA, Koomen M, Hendrick RE, Tardivon A, Toledano A (2012) Dual-energy contrast-enhanced digital mammography: initial clinical results of a multireader, multicase study. Breast Cancer Res 14(3):R94
Tagliafico AS, Bignotti B, Rossi F, Signori A, Sormani MP, Vadora F, Calabrese M, Houssami N (2016) Diagnostic performance of contrast-enhanced spectral mammography: systematic review and meta-analysis. Breast 28:13–19
Fei G, Teresa W, Jing L, Bin Z, Lingxiang R, Desheng S, Bhavika P (2018) SD-CNN: a shallow-deep CNN for improved breast cancer diagnosis. Comput Med Imaging Graph 70:53–62
Asmaa I, Paul G, Ronnachai J, Abdelsamea Mohammed M, Mermel Craig H, Po HCC, Rakha Emad A (2020) Artificial intelligence in digital breast pathology: techniques and applications. Breast 49:267–273
Arnau O, Jordi F, Joan M, Elsa P, Josep P, Denton Erika RE, Reyer Z (2010) A review of automatic mass detection and segmentation in mammographic images. Med Image Anal 14(2):87–110
Stephanie R, Hossein A, Kevin S, Johan H (2018) Digital image analysis in breast pathology from image processing techniques to artificial intelligence. Transl Res 194:19–35
Le EPV, Wang Y, Huang Y, Hickman S, Gilbert FJ (2019) Artificial intelligence in breast imaging. Clin Radiol 74(5):357–366
Habib G, Kiryati N, Sklair-Levy M, Shalmon A, Neiman O, Weidenfeld R, Yagil Y, Konen E, Mayer A (2020) Automatic breast lesion classification by joint neural analysis of mammography and ultrasound. In: Multimodal learning for clinical decision support and clinical image-based procedures: 10th international workshop, ML-CDS 2020, and 9th international workshop, CLIP 2020, held in conjunction with MICCAI 2020, Lima, Peru, October 4–8, 2020, Proceedings, vol 12445, pp 125–135. Springer
Shaikh TA, Rashid A, Sufyan Beg MM (2020) Transfer learning privileged information fuels cad diagnosis of breast cancer. Mach Vis Appl 31(1):1–23
Mateos MJ, Gastelum A, Márquez J, Brandan ME (2016). Texture analysis of contrast-enhanced digital mammography (CEDM) images. In: Lecture Notes in Computer Science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), vol 9699. Springer, pp 585–592
Shaked P, Nahum K, Gali Z-M, Miri S-L, Eli K, Arnaldo M (2019) Classification of contrast-enhanced spectral mammography (CESM) images. Int J Comput Assist Radiol Surg 14(2):249–257
Laura L, Menell Jennifer H (2002) Breast imaging reporting and data system (BI-RADS). Radiol Clin North Am 40(3):409–430
Fanizzi A, Losurdo L, Basile TMA, Bellotti R, Bottigli U, Delogu P, Diacono D, Didonna V, Fausto A, Lombardi A, Lorusso V, Massafra R, Tangaro S, Forgia DL (2019) Fully automated support system for diagnosis of breast cancer in contrast-enhanced spectral mammography images. J Clin Med 8(6):891
Liliana L, Annarita F, Basile Teresa Maria A, Roberto B, La Forgia D (2019) Radiomics analysis on contrast-enhanced spectral mammography images for breast cancer diagnosis: a pilot study. Entropy 21(11):1110
Daniele LF, Annarita F, Francesco C, Roberto B, Vittorio D, Vito L, Marco M, Raffaella M, Pasquale T, Sabina T, Michele T, Maria P, Alfredo Z (2020) Radiomic analysis in contrast-enhanced spectral mammography for predicting breast cancer histological outcome. Diagnostics 10:09
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Kaiming H, Xiangyu Z, Shaoqing R, Jian S (2016). Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Jie H, Li S, Gang S (2018). Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Saining X, Ross G, Piotr D, Zhuowen T, Kaiming H ( 2017). Aggregated residual transformations for deep neural networks. In: Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, vol 2017, pp 5987–5995
Zagoruyko S, Komodakis N (2016). Wide residual networks. In: British machine vision conference 2016, BMVC 2016, pp 87.1–87.12
Gao S, Cheng M-M, Zhao K, Zhang X-Y, Yang M-H, Torr PHS (2019). Res2net: a new multi-scale backbone architecture. IEEE transactions on pattern analysis and machine intelligence, pp :1–1, 08
Peng Qi, Wei Zhou, Jizhong Han (2017) A method for stochastic L-BFGS optimization, vol 434, pp 156–160
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2017). Grad-cam: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE international conference on computer vision, pp 618–626
Acknowledgements
This research was supported by Projects funded by National Natural Science Foundation of China Grant Numbers 81871508 and 61773246, the Major Program of Shandong Province Natural Science Foundation Grant Number ZR2018ZB0419, and the Taishan Scholar Program of Shandong Province of China grant number TSHW201502038.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Song, J., Zheng, Y., Zakir Ullah, M. et al. Multiview multimodal network for breast cancer diagnosis in contrast-enhanced spectral mammography images. Int J CARS 16, 979–988 (2021). https://doi.org/10.1007/s11548-021-02391-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02391-4