Abstract
Purpose
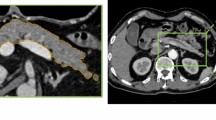
Pancreatic duct dilation can be considered an early sign of pancreatic ductal adenocarcinoma (PDAC). However, there is little existing research focused on dilated pancreatic duct segmentation as a potential screening tool for people without PDAC. Dilated pancreatic duct segmentation is difficult due to the lack of readily available labeled data and strong voxel imbalance between the pancreatic duct region and other regions. To overcome these challenges, we propose a two-step approach for dilated pancreatic duct segmentation from abdominal computed tomography (CT) volumes using fully convolutional networks (FCNs).
Methods
Our framework segments the pancreatic duct in a cascaded manner. The pancreatic duct occupies a tiny portion of abdominal CT volumes. Therefore, to concentrate on the pancreas regions, we use a public pancreas dataset to train an FCN to generate an ROI covering the pancreas and use a 3D U-Net-like FCN for coarse pancreas segmentation. To further improve the dilated pancreatic duct segmentation, we deploy a skip connection on each corresponding resolution level and an attention mechanism in the bottleneck layer. Moreover, we introduce a combined loss function based on Dice loss and Focal loss. Random data augmentation is adopted throughout the experiments to improve the generalizability of the model.
Results
We manually created a dilated pancreatic duct dataset with semi-automated annotation tools. Experimental results showed that our proposed framework is practical for dilated pancreatic duct segmentation. The average Dice score and sensitivity were 49.9% and 51.9%, respectively. These results show the potential of our approach as a clinical screening tool.
Conclusions
We investigate an automated framework for dilated pancreatic duct segmentation. The cascade strategy effectively improved the segmentation performance of the pancreatic duct. Our modifications to the FCNs together with random data augmentation and the proposed combined loss function facilitate automated segmentation.









Similar content being viewed by others
References
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O (2016) 3d u-net: learning dense volumetric segmentation from sparse annotation. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 424–432
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F (2013) The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digital Imag 26(6):1045–1057
Cubuk ED, Zoph B, Mane D, Vasudevan V, Le QV (2019) Autoaugment: learning augmentation strategies from data. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 113–123
Cubuk ED, Zoph B, Shlens J, Le QV (2020) Randaugment: practical automated data augmentation with a reduced search space. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops, pp 702–703
Edge MD, Hoteit M, Patel AP, Wang X, Baumgarten DA, Cai Q (2007) Clinical significance of main pancreatic duct dilation on computed tomography: single and double duct dilation. World J Gastroenterol WJG 13(11):1701
Guo C, Szemenyei M, Yi Y, Wang W, Chen B, Fan C (2020) Sa-unet: spatial attention u-net for retinal vessel segmentation. arXiv preprint arXiv:2004.03696
He K, Gkioxari G, Dollár P, Girshick R (2017) Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision, pp 2961–2969
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Hojjatoleslami S, Kittler J (1998) Region growing: a new approach. IEEE Trans Image Process 7(7):1079–1084
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning, pp 448–456
Kingma DP, Ba J (2015) Adam: a method for stochastic optimization. In: International conference for learning representations
Lin TY, Goyal P, Girshick R, He K, Dollár P (2017) Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision, pp 2980–2988
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Milletari F, Navab N, Ahmadi S (2016) V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV), pp 565–571 . https://doi.org/10.1109/3DV.2016.79
Mizrahi JD, Surana R, Valle JW, Shroff RT (2020) Pancreatic cancer. Lancet 395(10242):2008–2020
Nimura Y, Deguchi D, Kitasaka T, Mori K, Suenaga Y (2008) Pluto: a common platform for computer-aided diagnosis. Med Imag Technol 26(3):187
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D (2018) Attention u-net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Kopf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J, Chintala S (2019) Pytorch: an imperative style, high-performance deep learning library. In: Advances in neural information processing systems, pp 8026–8037
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Roth H, Oda M, Shimizu N, Oda H, Hayashi Y, Kitasaka T, Fujiwara M, Misawa K, Mori K (2018)Towards dense volumetric pancreas segmentation in ct using 3d fully convolutional networks. In: Medical imaging 2018: image processing, International society for optics and photonics, vol 10574, p 105740B
Roth HR, Farag A, Turkbey EB, Lu L, Liu J, Summers RM (2016) Data from pancreas-ct . https://doi.org/10.7937/K9/TCIA.2016.tNB1kqBU
Sata N, Kurihara K, Koizumi M, Tsukahara M, Yoshizawa K, Nagai H (2006) Ct virtual pancreatoscopy: a new method for diagnosing intraductal papillary mucinous neoplasm (IPMN) of the pancreas. Abdom Imag 31(3):326
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition
Tanaka S, Nakaizumi A, Ioka T, Oshikawa O, Uehara H, Nakao M, Yamamoto K, Ishikawa O, Ohigashi H, Kitamra T (2002) Main pancreatic duct dilatation: a sign of high risk for pancreatic cancer. Jpn J Clin Oncol 32(10):407–411
Xia Y, Xie L, Liu F, Zhu Z, Fishman EK, Yuille AL (2018) Bridging the gap between 2d and 3d organ segmentation with volumetric fusion net. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 445–453
Xia Y, Yu Q, Shen W, Zhou Y, Fishman EK, Yuille AL (2020) Detecting pancreatic ductal adenocarcinoma in multi-phase CT scans via alignment ensemble. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 285–295
Zhang L, Wang X, Yang D, Sanford T, Harmon S, Turkbey B, Wood BJ, Roth H, Myronenko A, Xu D, Xu Z (2020) Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans Med Imag
Zhou Y, Li Y, Zhang Z, Wang Y, Wang A, Fishman EK, Yuille AL, Park S (2019) Hyper-pairing network for multi-phase pancreatic ductal adenocarcinoma segmentation. Lect Notes Comput Sci Lect Notes Artif Intell Lect Notes Bioinform 11765:155–163. https://doi.org/10.1007/978-3-030-32245-8_18
Zhou Y, Xie L, Shen W, Wang Y, Fishman EK, Yuille, A.L (2017) A fixed-point model for pancreas segmentation in abdominal ct scans. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 693–701
Zhu Z, Xia Y, Xie L, Fishman EK, Yuille AL (2019) Multi-scale coarse-to-fine segmentation for screening pancreatic ductal adenocarcinoma. In: Shen D, Liu T, Peters TM, Staib LH, Essert C, Zhou S, Yap PT, Khan A (eds) Medical image computing and computer assisted intervention - MICCAI 2019. Springer International Publishing, Cham, pp 3–12
Acknowledgements
Part of this research was supported by MEXT/JSPS KAKENHI (894030, 17H00867, 21K19898) and the JSPS Bilateral Joint Research Project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the institutional review board of the Chiba Kensei Hospital.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shen, C., Roth, H.R., Hayashi, Y. et al. A cascaded fully convolutional network framework for dilated pancreatic duct segmentation. Int J CARS 17, 343–354 (2022). https://doi.org/10.1007/s11548-021-02530-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-021-02530-x