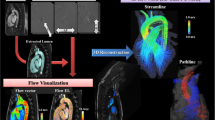
Abstract
Purpose
Communicating complex blood flow patterns generated from computational fluid dynamics (CFD) simulations to clinical audiences for the purposes of risk assessment or treatment planning is an ongoing challenge. While attempts have been made to develop new software tools for such clinical visualization of CFD data, these often overlook established medical imaging/visualization practice and data infrastructures. Here, leveraging the clinical ubiquity of the DICOM file format, we present techniques for the translation of CFD data to DICOM series, facilitating interactive visualization in standard radiological software.
Methods
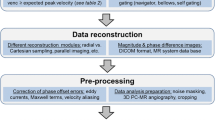
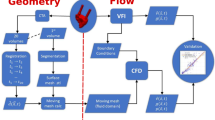
Unstructured CFD data (volumetric fields of velocity magnitude, Q-criterion, and pathlines) are resampled to structured grids. Novel raster-based techniques that simulate experimental optical blurring are presented for bringing simulated pathlines into structured image volumes. DICOM series are created by strategically encoding these data into the file’s PixelArray tag. Lumen surface information is also strategically encoded into a different range of pixel intensities, allowing hemodynamics and morphology to be co-visualized in a single volume using opacity-based rendering transfer functions.
Results
We show that 3D temporal CFD data represented as structured DICOM series can be rendered interactively in Horos, a widely-used medical imaging/radiology software. Our transfer function-based approach allows for representations of scalar isosurfaces, volumetric rendering, and tubular pathlines to be modified in real-time, resembling conventional unstructured visualizations. Careful selection of voxelization ROIs helps to ensure that data are kept lightweight for real-time rendering and minimal storage.
Conclusion
While our approach inherently sacrifices some of the advanced visualization capabilities of specialized software tools, we believe our closer consideration of standardization can help to facilitate meaningful clinical interaction. This work opens up possibilities for the complete integration of measured and simulated data in established radiological software environments and workflows from PACS storage to 3D/4D visualization.








Similar content being viewed by others
References
Caro CG (2009) Discovery of the role of wall shear in atherosclerosis. Arterioscler Thromb Vasc Biol 29(2):158–161. https://doi.org/10.1161/ATVBAHA.108.166736
Stankovic Z, Allen BD, Garcia J, Jarvis KB, Markl M (2014) 4D flow imaging with MRI. Cardiovasc Diagn Ther 4(2):173–192. https://doi.org/10.3978/j.issn.2223-3652.2014.01.02
Morris PD et al (2016) Computational fluid dynamics modelling in cardiovascular medicine. Heart 102(1):18–28. https://doi.org/10.1136/heartjnl-2015-308044
Steinman DA, Pereira VM (2019) How patient specific are patient-specific computational models of cerebral aneurysms? An overview of sources of error and variability. Neurosurg Focus 47(1):E14. https://doi.org/10.3171/2019.4.FOCUS19123
Levitt MR, Aliseda A, Fiorella D, Sadasivan C (2021) One way to get there. J Neurointerv Surg 13(5):401–402. https://doi.org/10.1136/neurintsurg-2021-017559
Pereira VM et al (2013) A DSA-based method using contrast-motion estimation for the assessment of the intra-aneurysmal flow changes induced by flow-diverter stents. AJNR Am J Neuroradiol 34(4):808–815. https://doi.org/10.3174/ajnr.A3322
Eulzer P, Meuschke M, Klingner C, Lawonn K (2021) Visualizing carotid blood flow simulations for stroke prevention. arXiv preprint arXiv:2104.02654
Chung B, Cebral JR (2015) CFD for evaluation and treatment planning of aneurysms: review of proposed clinical uses and their challenges. Ann Biomed Eng 43(1):122–138. https://doi.org/10.1007/s10439-014-1093-6
Kallmes DF (2012) Point: CFD—computational fluid dynamics or confounding factor dissemination. AJNR Am J Neuroradiol 33(3):395–396. https://doi.org/10.3174/ajnr.A2993
Gillmann C et al (2021) Ten open challenges in medical visualization. IEEE Comput Graph Appl 41(5):7–15. https://doi.org/10.1109/MCG.2021.3094858
Meuschke M, Preim B, Lawonn K (2021) Aneulysis—a system for the visual analysis of aneurysm data. Comput Graph 98:197–209. https://doi.org/10.1016/j.cag.2021.06.001
Xiang J et al (2016) AView: an image-based clinical computational tool for intracranial aneurysm flow visualization and clinical management. Ann Biomed Eng 44(4):1085–1096. https://doi.org/10.1007/s10439-015-1363-y
Wu J, Ho H, Hunter P, Liu P (2014) AneuSearch: a software prototype for intracranial aneurysm searching and clinical decision support. Int J Comput Assist Radiol Surg 9(6):997–1004. https://doi.org/10.1007/s11548-014-0996-x
Valen-Sendstad K, Steinman DA (2014) Mind the gap: impact of computational fluid dynamics solution strategy on prediction of intracranial aneurysm hemodynamics and rupture status indicators. AJNR Am J Neuroradiol 35(3):536–543. https://doi.org/10.3174/ajnr.A3793
Pereira VM et al (2020) Torrents of torment: turbulence as a mechanism of pulsatile tinnitus secondary to venous stenosis revealed by high-fidelity computational fluid dynamics. J Neurointerv Surg. https://doi.org/10.1136/neurintsurg-2020-016636
Natarajan T, MacDonald DE, Najafi M, Coppin PW, Steinman DA (2020) Spectral decomposition and illustration-inspired visualisation of highly disturbed cerebrovascular blood flow dynamics. Comput Methods Biomech Biomed Eng Imaging Vis 8(2):182–193. https://doi.org/10.1080/21681163.2019.1647461
Ahrens J, Geveci B, Law C (2005) Paraview: an end-user tool for large data visualization. The visualization handbook 717(8)
Smistad E, Bozorgi M, Lindseth F (2015) FAST: framework for heterogeneous medical image computing and visualization. Int J Comput Assist Radiol Surg 10(11):1811–1822. https://doi.org/10.1007/s11548-015-1158-5
Scherer S, Treichel T, Ritter N, Triebel G, Drossel WG, Burgert O (2011) Surgical stent planning: simulation parameter study for models based on DICOM standards. Int J Comput Assist Radiol Surg 6(3):319–327. https://doi.org/10.1007/s11548-010-0511-y
Cebral JR, Mut F, Weir J, Putman CM (2011) Association of hemodynamic characteristics and cerebral aneurysm rupture. AJNR Am J Neuroradiol 32(2):264–270. https://doi.org/10.3174/ajnr.A2274
Valen-Sendstad K, Piccinelli M, Steinman DA (2014) High-resolution computational fluid dynamics detects flow instabilities in the carotid siphon: implications for aneurysm initiation and rupture? J Biomech 47(12):3210–3216. https://doi.org/10.1016/j.jbiomech.2014.04.018
Steinman DA (2000) Simulated pathline visualization of computed periodic blood flow patterns. J Biomech 33(5):623–628. https://doi.org/10.1016/s0021-9290(99)00205-5
Westermann R (2001) The rendering of unstructured grids revisited. In: Data visualization 2001, pp 65–74. https://doi.org/10.1007/978-3-7091-6215-6_8
Purview. Horos. [Online]. https://horosproject.org. Accessed 01 Nov 2021
Schroeder W, Martin K, Lorensen B (2006) The visualization toolkit, 4th edn. Kitware
Kaufman A, Shimony E (1987) 3D scan-conversion algorithms for voxel-based graphics. In: Proceedings of the 1986 workshop on Interactive 3D graphics, pp 45–75. https://doi.org/10.1145/319120.319126.
Mason D et al. pydicom: an open source DICOM library. [Online]. https://github.com/pydicom/pydicom
Mori S, van Zijl PCM (2002) Fiber tracking: principles and strategies—a technical review. NMR Biomed 15(7–8):468–480. https://doi.org/10.1002/nbm.781
Cebral JR, Meng H (2012) Counterpoint: realizing the clinical utility of computational fluid dynamics—closing the gap. Am J Neuroradiol 33(3):396–398. https://doi.org/10.3174/ajnr.A2994
Funding
This work was supported by a grant to DAS from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04649). LT was also supported by a Barbara and Frank Milligan Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Temor, L., Cancelliere, N.M., MacDonald, D.E. et al. Integrating computational fluid dynamics data into medical image visualization workflows via DICOM. Int J CARS 17, 1143–1154 (2022). https://doi.org/10.1007/s11548-022-02613-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02613-3