Abstract
Purpose
Radiotherapy is one of most treatments for tumors. To accurately control the radiation dose distribution and lessen the radiation damage to normal tissues and organs in radiotherapy, it is essential to delineate organs at risk (OARs) precisely. However, manual delineating and some traditional methods are labor-intensive and time-consuming. There is an urgent need for fast and precise segmentation methods in radiotherapy.
Methods
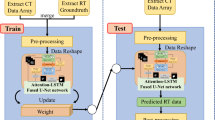
This paper proposes a fully automatic segmentation method based on the 3D U-Net for multi-organ in head and neck. It introduces squeeze-and-attention blocks to gather multi-scale context information and the receptive field block to balance the performance between large-sized and small-sized organs. Furthermore, it is trained by the marginal and exclusion loss function in a partially supervised learning mode.
Results
We evaluated the model with dice similarity coefficient (DSC), 95% Hausdorff distance (95HD) and inference time. Its average DSC is 0.829, which is 4.5%, 3.2%, and 2.4% higher than AnatomyNet’s, nnU-net’s, and FocusNet’s, respectively, and its average 95HD is 2.19. Moreover, its inference time and parameters are 63% and 60% less than FocusNetv2’s.
Conclusion
For the segmentation of OARs in head and neck, our model is more accurate than AnatomyNet, faster than FocusNetv2, and better balances between segmentation accuracy and inference time. It demonstrates that our method is more applicable for clinical treatment.





Similar content being viewed by others
References
Jemal A, Bray F, Center MM (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. https://doi.org/10.3322/caac.20107
Çiçek Ö, Abdulkadir A, Lienkamp S.S., Brox T, Ronneberger O (2016) 3d u-net: Learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) Int. Conf. on Medical Image Computing and Computer-assisted Intervention, pp. 424–432. Springer, Cham . https://doi.org/10.1007/978-3-319-46723-8_49. https://link.springer.com/chapter/10.1007%2F978-3-319-46723-8_49
Zhong Z, Lin Z.Q., Bidart R, Hu X, Daya I.B., Li Z, Zheng W.S., Li J, Wong A (2020) Squeeze-and-attention networks for semantic segmentation. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 13062–13071 . https://doi.org/10.1109/CVPR42600.2020.01308
Liu S, Huang D, Wang Y (2018) Receptive field block net for accurate and fast object detection. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) Computer Vision –ECCV 2018, pp. 404–419. Springer, Cham . https://doi.org/10.1007/978-3-030-01252-6_24
Shi G, Xiao L, Chen Y, Zhou S.K. (2021) Marginal loss and exclusion loss for partially supervised multi-organ segmentation. Medical Image Analysis 70, 101979 . https://doi.org/10.1016/j.media.2021.101979
Ren X, Lei X, Dong N, Shao Y, Zhang H, Shen D, Qian W (2018) Interleaved 3d-cnns for joint segmentation of small-volume structures in head and neck ct images. Medical Physics 45(5), 2063–2075 . https://doi.org/10.1002/mp.12837
Vrtovec T, Močnik D, Strojan P, Pernuš F, Ibragimov B (2020) Auto-segmentation of organs at risk for head and neck radiotherapy planning: From atlas-based to deep learning methods. Medical Physics 47(9), 929–950 . https://doi.org/10.1002/mp.14320
Gou S, Tong N, Qi S, Yang S, Chin R, Sheng K (2020) Self-channel-and-spatial-attention neural network for automated multi-organ segmentation on head and neck CT images. Physics in Medicine & Biology 65(24), 245034 . https://doi.org/10.1088/1361-6560/ab79c3
Zhu W, Huang Y, Liang Z, Chen X, Yong L, Zhen Q, Nan D, Wei F, Xie X (2018) Anatomynet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Medical Physics 46(2), 576–589 . https://doi.org/10.1088/1361-6560/abd953
Hu J, Shen L, Albanie S, Sun G, Wu E (2020) Squeeze-and-excitation networks. IEEE Transactions on Pattern Analysis and Machine Intelligence 42(8), 2011–2023 . https://doi.org/10.1109/TPAMI.2019.2913372
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 39(4), 640–651 . https://doi.org/10.1109/TPAMI.2016.2572683
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2020) Focal loss for dense object detection. IEEE Transactions on Pattern Analysis and Machine Intelligence 42(2):318–327. https://doi.org/10.1109/TPAMI.2018.2858826
Tappeiner E, Pröll S, Hönig M, Raudaschl P.F., Zaffino P, Spadea M.F., Sharp G.C., Schubert R, Fritscher K (2019) Multi-organ segmentation of the head and neck area: an efficient hierarchical neural networks approach. International Journal of Computer Assisted Radiology and Surgery 14, 745–759 . https://doi.org/10.1007/s11548-019-01922-4
Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, Yang Y, Zhang J, Tao C, Li H (2019) FocusNet: Imbalanced Large and Small Organ Segmentation with an End-to-End Deep Neural Network for Head and Neck CT Images. https://arxiv.org/abs/1907.12056v1
Gao Y, Huang R, Yang Y, Zhang J, Shao K, Tao C, Chen Y, Metaxas D.N., Li H, Chen M (2021) Focusnetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck ct images. Medical Image Analysis 67, 101831. https://doi.org/10.1016/j.media.2020.101831
Liu, Y., Lei, Y., Fu, Y., Wang, T., Zhou, J., Jiang, X., McDonald, M., Beitler, J.J., Curran, W.J., Liu, T., Yang, X.: Head and neck multi–organ auto-segmentation on ct images aided by synthetic mri. Medical Physics 47(9), 4294–4302 (2020). https://doi.org/10.1002/mp.14378
Xu, X., Chen, J., Zhang, H., Han, G.: Dual pyramid network for salient object detection. Neurocomputing 375, 113–123 (2020). https://doi.org/10.1016/j.neucom.2019.09.077
Dai, X., Lei, Y., Wang, T., Dhabaan, A.H., McDonald, M., Beitler, J.J., Curran, W.J., Zhou, J., Liu, T., Yang, X.: Head-and-neck organs-at-risk auto-delineation using dual pyramid networks for CBCT-guided adaptive radiotherapy. Physics in Medicine & Biology 66(4), 045021 (2021). https://doi.org/10.1088/1361-6560/abd953
Raudaschl, P., Zaffino, P., Sharp, G., Spadea, M., Chen, A., Dawant, B.M., Albrecht, T., Gass, T., Langguth, C., Lüthi, M., Jung, F., Knapp, O., Wesarg, S., Mannion-Haworth, R., Bowes, M., Ashman, A., Guillard, G., Brett, A., Vincent, G., Orbes-Arteaga, M., Cárdenas-Pen̄a, D., Castellanos-Dominguez, G., Aghdasi, N., Li, Y., Berens, A., Hannaford, B., Schubert, R., Fritscher, K.D.: Evaluation of segmentation methods on head and neck ct: Auto-segmentation challenge 2015. Medical Physics 44(5), 2020–2036 (2017). https://doi.org/10.1002/mp.12197
Clark, Vendt, Smith, Freymann, Kirby, Koppel, Moore, Phillips, Maffitt, and, P. (2013) The cancer imaging archive (tcia): Maintaining and operating a public information repository. Digit Imaging 26, 1045–1057 . https://doi.org/10.1007/s10278-013-9622-7
Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts H, Khaouam N, Nguyen-Tan PF, Wang CS, Sultanem K (2017) Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Scientific Reports 7(1):10117. https://doi.org/10.1038/s41598-017-10371-5
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2818–2826 . https://doi.org/10.1109/CVPR.2016.308
Chen L.-C., Papandreou G, Kokkinos I, Murphy K, Yuille A.L. (2018) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence 40(4), 834–848 . https://doi.org/10.1109/TPAMI.2017.2699184
Isensee F, Jaeger P.F., Kohl S.A.A., Petersen J, Maier-Hein K.H. (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 18, 203–211 . https://doi.org/10.1038/s41592-020-01008-z
Chen A, Dawant B (2016) A multi-atlas approach for the automatic segmentation of multiple structures in head and neck ct images . https://doi.org/10.54294/hk5bjs
Albrecht T, Gass T, Langguth C, Lüthi M (2015) Multi atlas segmentation with active shape model refinement for multi-organ segmentation in head and neck cancer radiotherapy planning . https://doi.org/10.54294/kmcunc
Funding
We would like to thank the financial supports from National Natural Science Foundation of China (62175156, 61976140, 61675134, 81827807), Shanghai Committee of Science and Technology (1944190 5800), and Wenzhou Medical University Key Laboratory Open Project (K181002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
For the retrospective studies formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hongdong, M., Guogang, C., Shu, Z. et al. Multi-scale organs image segmentation method improved by squeeze-and-attention based on partially supervised learning. Int J CARS 17, 1135–1142 (2022). https://doi.org/10.1007/s11548-022-02632-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02632-0