Abstract
Purpose
Speed and accuracy are two critical factors in dose calculation for radiotherapy. Analytical Anisotropic Algorithm (AAA) is a rapid dose calculation algorithm but has dose errors in tissue margin area. Acuros XB (AXB) has high accuracy but takes long time to calculate. To improve the dose accuracy on the tissue margin area for AAA, we proposed a novel deep learning-based dose accuracy improvement method using Margin-Net combined with Margin-Loss.
Methods
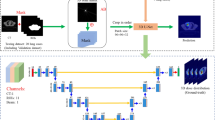
A novel model ‘Margin-Net’ was designed with a Margin Attention Mechanism to generate special margin-related features. Margin-Loss was introduced to consider the dose errors and dose gradients in tissues margin area. Ninety-five VMAT cervical cancer cases with paired AAA and AXB dose were enrolled in our study: 76 cases for training and 19 cases for testing. Tissues Margin Masks were generated from RT contours with 6 mm extension. Tissues Margin Mask, AAA dose and CTs were input data; AXB dose was used as reference dose for model training and evaluation. Comparison experiments were performed to evaluated effectiveness of Margin-Net and Margin-Loss.
Results
Compared to AXB dose, the 3D gamma passing rate (1%/1 mm, 10% threshold) for 19 test cases 95.75 ± 1.05% using Margin-Net with Margin-Loss, which was significantly higher than the original AAA dose (73.64 ± 3.46%). The passing rate reduced to 94.07 ± 1.16% without Margin-Loss and 87.3 ± 1.18% if Margin-Net key structure ‘MAM’ was also removed.
Conclusion
The proposed novel tissues margin-based dose conversion method can significantly improve the dose accuracy of Analytical Anisotropic Algorithm to be comparable to AXB algorithm. It can potentially improve the efficiency of treatment planning process with low demanding of computation resources.




Similar content being viewed by others
References
Wulf J, Baier K, Mueller G, Flentje MP (2005) Dose-response in stereotactic irradiation of lung tumors. Radiother Oncol 77(1):83–87. https://doi.org/10.1016/j.radonc.2005.09.003
Partridge M, Ramos M, Sardaro A, Brada M (2011) Dose escalation for non-small cell lung cancer: analysis and modelling of published literature. Radiother Oncol 99(1):6–11. https://doi.org/10.1016/j.radonc.2011.02.014
Sievinen J, Ulmer W, Kaissl W (2005) AAA photon dose calculation model in Eclipse. Palo Alto (CA): Varian Medical Systems. 118:2894.
Van Esch A, Tillikainen L, Pyykkonen J, Tenhunen M, Helminen H, Siljamäki S, Alakuijala J, Paiusco M, Iori M, Huyskens DP (2006) Testing of the analytical anisotropic algorithm for photon dose calculation. Med Phys 33(11):4130–4148. https://doi.org/10.1118/1.2358333
Robinson D (2008) Inhomogeneity correction and the analytic anisotropic algorithm. J Appl Clin Med Phys 9(2):112–122. https://doi.org/10.1120/jacmp.v9i2.2786
Bragg CM, Conway J (2006) Dosimetric verification of the anisotropic analytical algorithm for radiotherapy treatment planning. Radiother Oncol 81(3):315–323. https://doi.org/10.1016/j.radonc.2006.10.020
Xing Y, Zhang Y, Nguyen D, Lin MH, Lu W, Jiang S (2020) Boosting radiotherapy dose calculation accuracy with deep learning. J Appl Clin Med Phys 21(8):149–159. https://doi.org/10.1002/acm2.12937
Ma C-M, Li J, Pawlicki T, Jiang S, Deng J, Lee M, Koumrian T, Luxton M, Brain S (2002) A Monte Carlo dose calculation tool for radiotherapy treatment planning. Phys Med Biol 47(10):1671. https://doi.org/10.1088/0031-9155/47/10/305
Failla GA, Wareing T, Archambault Y, Thompson S (2010) Acuros XB advanced dose calculation for the Eclipse treatment planning system. Varian Med Syst 20:18
Kan MW, Yu PK, Leung LH (2013) A review on the use of grid-based Boltzmann equation solvers for dose calculation in external photon beam treatment planning. BioMed Res Int. https://doi.org/10.1155/2013/692874.
Rana S, Rogers K (2013) Dosimetric evaluation of Acuros XB dose calculation algorithm with measurements in predicting doses beyond different air gap thickness for smaller and larger field sizes. J Med Phys/Assoc Med Physicists India 38(1):9. https://doi.org/10.4103/0971-6203.106600
Dong P, Xing L (2020) Deep DoseNet: a deep neural network for accurate dosimetric transformation between different spatial resolutions and/or different dose calculation algorithms for precision radiation therapy. Phys Med Biol 65(3):035010. https://doi.org/10.1088/1361-6560/ab652d
Wu C, Nguyen D, Xing Y, Montero A, Schuemannn J, Shang H, Pu Y, Jiang S (2021) Improving proton dose calculation accuracy by using deep learning. Mach Learn Sci Technol 2(1):015017. https://doi.org/10.1088/2632-2153/abb6d5
Sumida I, Magome T, Das IJ, Yamaguchi H, Kizaki H, Aboshi K, Yamaguchi H, Seo Y, Isohashi F, Ogawa K (2020) A convolution neural network for higher resolution dose prediction in prostate volumetric modulated arc therapy. Phys Med 72:88–95. https://doi.org/10.1016/j.ejmp.2020.03.023
Woo S, Park J, Lee J-Y, Kweon IS (2018) Cbam: Convolutional block attention module. Proceedings of the European conference on computer vision (ECCV) 2018. https://doi.org/10.48550/arXiv.1807.06521.
Zhang Z (2018) Improved adam optimizer for deep neural networks. In: 2018 IEEE/ACM 26th International Symposium on Quality of Service (IWQoS), pp 1–2. https://doi.org/10.1109/IWQoS.2018.8624183.
Nguyen D, Jia X, Sher D, Lin MH, Iqbal Z, Liu H, Jiang S (2019) 3D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture. Phys Med Biol 64(6):065020. https://doi.org/10.1088/1361-6560/ab039b
Pieper S, Halle M, Kikinis R (2004) 3D Slicer. Paper presented at: 2004 2nd IEEE international symposium on biomedical imaging: nano to macro (IEEE Cat No. 04EX821) 2004. https://doi.org/10.1109/ISBI.2004.1398617.
Kry SF, Feygelman V, Balter P, Knöös T, Charlie Ma C-M, Snyder M, Tonner B, Vassiliev ONAAPM (2020) Task Group 329: Reference dose specification for dose calculations: Dose-to-water or dose-to-muscle? Med Phys 47:e52–e64. https://doi.org/10.1002/mp.13995
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. https://doi.org/10.1007/978-3-319-24574-4_28.
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition. https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. https://doi.org/10.48550/arXiv.1608.06993.
Liu Y, Chen Z, Wang J, Wang X, Qu B, Ma L, Zhao W, Zhang G, Xu S (2021) Dose prediction using a three-dimensional convolutional neural network for nasopharyngeal carcinoma with tomotherapy. FrontiOncol, p 11. https://doi.org/10.3389/fonc.2021.752007.
Acknowledgements
We sincerely thank the editor and all reviewers for their very insightful comments and constructive suggestions, which really helped us improve the quality of this manuscript.
Funding
Funding Program: The China National Key R&D Program during the 13th Five-year Plan Period (Grant Nos. 2016YFC0105206, 2016YFC0105207).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There was no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, B., Liu, Y., Chen, Z. et al. Tissues margin-based analytical anisotropic algorithm boosting method via deep learning attention mechanism with cervical cancer. Int J CARS 18, 953–959 (2023). https://doi.org/10.1007/s11548-022-02801-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02801-1