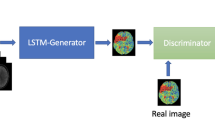
Abstract
Purpose
Multiple medical imaging modalities are used for clinical follow-up ischemic stroke analysis. Mixed-modality datasets are challenging, both for clinical rating purposes and for training machine learning models. While image-to-image translation methods have been applied to harmonize stroke patient images to a single modality, they have only been used for paired data so far. In the more common unpaired scenario, the standard cycle-consistent generative adversarial network (CycleGAN) method is not able to translate the stroke lesions properly. Thus, the aim of this work was to develop and evaluate a novel image-to-image translation regularization approach for unpaired 3D follow-up stroke patient datasets.
Methods
A modified CycleGAN was used to translate images between 238 non-contrast computed tomography (NCCT) and 244 fluid-attenuated inversion recovery (FLAIR) MRI datasets, two of the most relevant follow-up modalities in clinical practice. We introduced an additional attention-guided mechanism to encourage an improved translation of the lesion and a gradient-consistency loss to preserve structural brain morphology.
Results
The proposed modifications were able to preserve the overall quality provided by the CycleGAN translation. This was confirmed by the FID score and gradient correlation results. Furthermore, the lesion preservation was significantly improved compared to a standard CycleGAN. This was evaluated for location and volume with segmentation models, which were trained on real datasets and applied to the translated test images. Here, the Dice score coefficient resulted in 0.81 and 0.62 for datasets translated to FLAIR and NCCT, respectively, compared to 0.57 and 0.50 for the corresponding datasets translated using a standard CycleGAN. Finally, an analysis of the distribution of mean lesion intensities showed substantial improvements.
Conclusion
The results of this work show that the proposed image-to-image translation method is effective at preserving stroke lesions in unpaired modality translation, supporting its potential as a tool for stroke image analysis in real-life scenarios.






Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Fiehler J, Thomalla G, Bernhardt M, Kniep H, Berlis A, Dorn F, Eckert B, Kemmling A, Langner S, Remonda L, Reith W, Rohde S, Möhlenbruch M, Bendszus M, Forkert ND, Gellissen S (2019) ERASER: a thrombectomy study with predictive analytics end point. Stroke 50(5):1275–1278. https://doi.org/10.1161/STROKEAHA.119.024858
Makkat S, Vandevenne JE, Verswijvel G, Ijsewijn T, Grieten M, Palmers Y, De Schepper AM, Parizel PM (2002) Signs of acute stroke seen on fluid-attenuated inversion recovery MR imaging. Am J Roentgenol 179(1):237–243
Peultier A-C, Redekop WK, Dippel DW, Bereczki D, Si-Mohamed S, Douek PC, Severens JL (2019) What stroke image do we want? European survey on acute stroke imaging and revascularisation treatment. Health Policy Technol 8(3):261–267. https://doi.org/10.1016/j.hlpt.2019.08.005
Clèrigues A, Valverde S, Bernal J, Freixenet J, Oliver A, Lladó X (2020) Acute and sub-acute stroke lesion segmentation from multimodal MRI. Comput Methods Progr Biomed 194:105521. https://doi.org/10.1016/j.cmpb.2020.105521
Gillmann C, Peter L, Schmidt C, Saur D, Scheuermann G (2021) Visualizing multimodal deep learning for lesion prediction. IEEE Computer springer nature 2021 LATEX template 14 Lesion-preserving image-to-image translation graphics and applications 41(5), 90–98. https://doi.org/10.1109/MCG.2021.3099881
Lo Vercio L, Amador K, Bannister J, Crites S, Gutierrez A, MacDonald ME, Moore J, Mouches P, Rajasheka D, Schimert S, Subbanna N, Tuladhar A, Wang N, Wilms M, Winder A, Forkert ND (2020) Supervised machine learning tools: a tutorial for clinicians. J Neural Eng 17(6):062001. https://doi.org/10.1088/1741-2552/abbff2
MacEachern SJ, Forkert ND (2021) Machine learning for precision medicine. Genome 64(4):416–425. https://doi.org/10.1139/gen-2020-0131
Zhu J-Y, Park T, Isola P, Efros AA (2017) Unpaired image-to-image translation using cycle-consistent adversarial networks. In: Proceedings of the IEEE international conference on computer vision. pp. 2223–2232
Hiasa Y, Otak Y, Takao M, Matsuoka T, Takashima K, Carass A, Prince J, Sugano N, Sato Y (2018) Cross-modality image synthesis from unpaired data using CycleGAN. In: International workshop on simulation and synthesis in medical imaging. Springer, pp. 31–41. https://doi.org/10.1007/978-3-030-00536-84
Cohen JP, Luck M, Honari S (2018) Distribution matching losses can hallucinate features in medical image translation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp. 529–536. https://doi.org/10.1007/978-3-030-00928-160
Alami Mejjati Y, Richardt C, Tompkin J, Cosker D, Kim KI (2018) Unsupervised attention–guided image-to-image translation. Advances in neural information processing systems, 31
Emami H, DongM, Glide–Hurst, CK. (2020) Attention-guided generative adversarial network to address atypical anatomy in synthetic CT generation. In: 2020 IEEE 21st International conference on information reuse and integration for data science (IRI), pp. 188–193 IEEE https://doi.org/10.1109/IRI49571.2020.00034
Abu-Srhan A, Almallahi I, Abushariah MA, Mahafza W, Al-Kadi OS (2021) Paired-unpaired unsupervised attention guided GAN with transfer learning for bidirectional brain MR-CT synthesis. Comput Biol Med 136:104763. https://doi.org/10.1016/j.compbiomed.2021.104763
Gutierrez A, Tuladhar A, Rajashekar D, Forkert ND (2022) Lesion-preserving unpaired image-to-image translation between MRI and CT from ischemic stroke patients. In: Medical imaging 2022: computer-aided diagnosis, vol. 12033, pp. 308–314 https://doi.org/10.1117/12.2613203
Demchuk A, Goyal M, Menon B, Eesa M, Ryckborst K, Kamal N, Patil S, Mishra S, Almekhlafi M, Randhawa P, Roy D, Willinsky R, Montanera W, Silver F, Shuaib A, Rempel J, Jovin T, Frei D, Sapkota B, Hill M (2015) Endovascular treatment for small core and anterior circulation proximal occlusion with emphasis on minimizing CT to recanalization times (ESCAPE) trial: methodology. Int J Stroke 10(3):429–438. https://doi.org/10.1111/ijs.12424
Cheng B, Forkert ND, Zavaglia M, Hilgetag C, Golsari A, Siemonsen S, Fiehler J, Pedraza S, Puig J, Cho T-H, Alawneh J, Baron J-C, Ostergaard L, Gerloff C, Thomalla G (2014) Influence of stroke infarct location on functional outcome measured by the modified rankin scale. Stroke 45(6):1695–1702. https://doi.org/10.1161/STROKEAHA.114.005152
Menon B, Al-Ajlan F, Najm M, Puig J, Castellanos M, Dowlatshahi D, Calleja A, Sohn S-I, Ahn SH, Poppe A, Mikulík R, Asdaghi N, Field T, Jin A, Asil T, Boulanger J-M, Smith E, Coutts S, Barber P, Demchuk A (2018) Association of clinical, imaging, and thrombus characteristics with recanalization of visible intracranial occlusion in patients with acute ischemic stroke. JAMA 320(10):1017–1026. https://doi.org/10.1001/jama.2018.12498
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54(3):2033–2044. https://doi.org/10.1016/j.neuroimage.2010.09.025
Muschelli J, Ullman NL, Mould WA, Vespa P, Hanley DF, Crainiceanu CM (2015) Validated automatic brain extraction of head CT images. Neuroimage 114:379–385. https://doi.org/10.1016/j.neuroimage.2015.03.074
Rajashekar D, Wilms M, MacDonald ME, Ehrhardt J, Mouches P, Frayne R, Hill MD, Forkert ND (2020) High-resolution T2-FLAIR and non-contrast CT brain atlas of the elderly. Sci Data 7(1):1–7. https://doi.org/10.1038/s41597-020-0379-9
Ehrhardt J, Säring D, Handels H (2007) Structure-preserving interpolation of temporal and spatial image sequences using an optical flow-based method. Methods Inf Med 46(03):300–307. https://doi.org/10.1160/ME9047
Heusel M, Ramsauer H, Unterthiner T, Nessler B, & Hochreiter S (2017) Gans trained by a two time-scale update rule converge to a local nash equilibrium. Advances in neural information processing systems, 30
Zhang Y, Liu S, Li C, Wang J (2021) Rethinking the dice loss for deep learning lesion segmentation in medical images. J Shanghai Jiaotong Univ (Sci) 26:93–102. https://doi.org/10.1007/s12204-021-2264-x
Thiyagarajan SK, Murugan K (2021) A systematic review on techniques adapted for segmentation and classification of ischemic stroke lesions from brain MR images. Wireless Pers Commun 118(2):1225–1244. https://doi.org/10.1007/s11277-021-08069-z
Broocks G, Leischner H, Hanning U, Flottmann F, Faizy T, Schön G, Sporns P, Thomalla G, Kamalian S, Lev M, Fiehler J, Kemmling A (2020) Lesion age imaging in acute stroke: water uptake in CT versus DWI-FLAIR mismatch. Ann Neurol 88(6):1144–1152
Qazi E, Al-Ajlan F, Mahajan A, Sohn S-I, Mishra SChang H, Najm M, d’Esterre C, Demchuk A, Goyal M, Lee T, Hill M, Menon B (2016) Non-contrast CT in place of MRI mismatch in the imaging triage of acute ischemic stroke patients. Med Res Arch 4(6)
Funding
This work was supported by the Canada Research Chairs program, the River Fund at Calgary Foundation, Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canadian Open Neuroscience Platform (CONP).
Author information
Authors and Affiliations
Contributions
AG contributed to the study conception. AG, AT, MW, DR, and NDF contributed to the study design. Experiments were performed by AG. The data was collected by MDH, AD, MG, and JF. The first draft of the manuscript was written by AG and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Acquisition of the datasets was approved by the respective local ethics board at each contributing site.
Informed consent
All datasets were made available for this secondary study after complete anonymization.
Code availability
The source code used to compute the experiments in this study will be made available on https://github.com/Alexhal9000/lesion-preserving-cyclegan
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gutierrez, A., Tuladhar, A., Wilms, M. et al. Lesion-preserving unpaired image-to-image translation between MRI and CT from ischemic stroke patients. Int J CARS 18, 827–836 (2023). https://doi.org/10.1007/s11548-022-02828-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02828-4