Abstract
Purpose
Medical image registration is of great importance in clinical medicine. However, medical image registration algorithms are still in the development stage due to the challenges posed by the related complex physiological structures. The objective of this study was to design a 3D medical image registration algorithm that satisfies the need for high accuracy and speed of complex physiological structures.
Methods
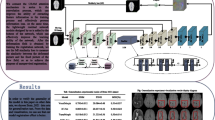
We present a new unsupervised learning algorithm, “DIT-IVNet,” for 3D medical image registration. Unlike the more popular convolution-based U-shaped registration network architectures like VoxelMorph, DIT-IVNet uses a combined convolution and transformer network architecture. To better extract image information features and reduce the heavy training parameters, we improved the 2D_Depatch module to a 3D_Depatch module, thus replacing the patch embedding in the original Vision Transformer which adaptively performs patch embedding based on 3D image structure information. We also designed inception blocks in the down-sampling part of the network to help coordinate feature learning from images to different scales.
Results
Dice score, Negative Jacobian determinant, Hausdorff distance, and Structural Similarity evaluation metrics were used to evaluate the registration effects. The results showed that our proposed network had the best metric results compared with some state-of-the-art methods. Moreover, our network obtained the highest Dice score in the generalization experiments which indicated better generalizability of our model.
Conclusion
We proposed an unsupervised registration network and evaluated its performance in deformable medical image registration. The results of the evaluation metrics showed that the network structure outperformed state-of-the-art methods for the registration of brain datasets.








Similar content being viewed by others
References
Xiao H, Teng X, Liu C, Li T, Ren G, Yang R, Shen D, Cai J (2021) A review of deep learning-based three-dimensional medical image registration methods. Quant Imaging Med Surg 11:4895
Chen X, Diaz-Pinto A, Ravikumar N, Frangi AF (2021) Deep learning in medical image registration. Prog Biomed Eng 3:012003
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ANTS). Insight J 2:1–35
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2009) Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29:196–205
Gupta S, Gupta P, Verma VS (2021) Study on anatomical and functional medical image registration methods. Neurocomputing 452:534–548
Sokooti H, De Vos B, Berendsen F, Lelieveldt BP, Išgum I, Staring M (2017) Nonrigid image registration using multi-scale 3D convolutional neural networks. In: International conference on medical image computing and computer-assisted intervention, 232–239
Nagel H-H, Enkelmann W (1986) An investigation of smoothness constraints for the estimation of displacement vector fields from image sequences. IEEE Trans Pattern Anal 8:565–593. https://doi.org/10.1109/TPAMI.1986.4767833
Heinrich MP (2019) Closing the gap between deep and conventional image registration using probabilistic dense displacement networks. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, 50–58
Zhou Z-H (2018) A brief introduction to weakly supervised learning. Natl Sci Rev 5:44–53. https://doi.org/10.1093/nsr/nwx106
Dosovitskiy A, Fischer P, Ilg E, Hausser P, Hazirbas C, Golkov V, Van Der Smagt P, Cremers D, Brox T (2015) Flownet: learning optical flow with convolutional networks. In: Proceedings of the IEEE international conference on computer vision, 2758–2766
Yang X, Kwitt R, Styner M, Niethammer M (2017) Quicksilver: fast predictive image registration–a deep learning approach. Neuroimage 158:378–396
Krebs J, Mansi T, Delingette H, Zhang L, Ghesu FC, Miao S, Maier AK, Ayache N, Liao R, Kamen A (2017) Robust non-rigid registration through agent-based action learning. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, 344–352.
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) Voxelmorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38:1788–1800. https://doi.org/10.1109/TMI.2019.2897538
Kuang D, Schmah T. (2019) Faim–a convnet method for unsupervised 3d medical image registration. In: International Workshop on Machine Learning in Medical Imaging. Springer, Cham pp 646–654
Mok TC, Chung A. (2020) Fast symmetric diffeomorphic image registration with convolutional neural networks. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp 4644–4653
Chen J, He Y, Frey EC, Li Y, Du Y (2021) ViT-V-Net: Vision Transformer for Unsupervised Volumetric Medical Image Registration. Electrical Engineering and Systems Science. https://arxiv.org/abs/2014.06468.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I (2017) Attention is all you need. Adv Neural Inf Process Syst 30:5998–6008
Chen Z, Zhu Y, Zhao C, Hu G, Zeng W, Wang J, Tang M (2021) DPT: deformable patch-based transformer for visual recognition. In: Proceedings of the 29th ACM International Conference on Multimedia, 2899–2907
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 1–9.
Ota K, Oishi N, Ito K, Fukuyama H, Group S-JS (2014) A comparison of three brain atlases for MCI prediction. J Neurosci Methods 221:139–150
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L (2019) Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst 32:8026–8037
Willmott CJ, Matsuura K (2005) Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Clim Res 30:79–82
Kim B, Kim DH, Park SH, Kim J, Lee JG, Ye JC (2021) CycleMorph: cycle consistent unsupervised deformable image registration. Med Image Anal 71:102036
Bilic P, Christ PF, Vorontsov E, Chlebus G, Chen H, Dou Q, Fu C-W, Han X, Heng P-A (2019) The liver tumor segmentation benchmark (lits). arXiv preprint arXiv: 04056
Acknowledgements
The work was supported by the National Science Foundation for Young Scientists of China (Grant No.61806060), 2019-2021, the Natural Science Foundation of Heilongjiang Province (LH2019F024), China, 2019-2021; the Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515220140) the Youth Innovation Project of Sun Yat-sen University Cancer Center (QNYCPY32) and the Science Foundation of Guangzhou Xinhua University(2020YQYJ05).
Funding
The National Science Foundation for Young Scientists of China, N0.61806060, Liwei Deng, the Natural Science Foundation of Heilongjiang Province, LH2019F024, Liwei Deng, the Youth Innovation Project of Sun Yat-sen University Cancer Center, QNYCPY32, Xin Yang, Basic and Applied Basic Research Foundation of Guangdong Province, 2021A1515220140, Xin Yang.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Humans and animals participants
The LPBA40 dataset used during this study is a publicly available dataset from the Mark and Mary Stevens Institute for Neuroimaging and Informatics at the University of Southern California, [https://www.loni.usc.edu/research]. The LiTS-2017 dataset used in this study is a publicly available dataset from the IEEE International Symposium on Biomedical Imaging, [https://competitions.codalab.org/competitions/17094].
Informed consent
The images in the dataset used have been reused to obtain informed consent from all individual participants included in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, L., Zhi, Q., Huang, S. et al. A deformable patch-based transformer for 3D medical image registration. Int J CARS 18, 2295–2306 (2023). https://doi.org/10.1007/s11548-023-02860-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-023-02860-y