Abstract
Purpose
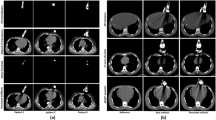
The strong metal artifacts produced by the electrode needle cause poor image quality, thus preventing physicians from observing the surgical situation during the puncture process. To address this issue, we propose a metal artifact reduction and visualization framework for CT-guided ablation therapy of liver tumors.
Methods
Our framework contains a metal artifact reduction model and an ablation therapy visualization model. A two-stage generative adversarial network is proposed to reduce the metal artifacts of intraoperative CT images and avoid image blurring. To visualize the puncture process, the axis and tip of the needle are localized, and then the needle is rebuilt in 3D space intraoperatively.
Results
Experiments show that our proposed metal artifact reduction method achieves higher SSIM (0.891) and PSNR (26.920) values than the state-of-the-art methods. The accuracy of ablation needle reconstruction is 2.76 mm average in needle tip localization and 1.64° average in needle axis localization.
Conclusion
We propose a novel metal artifact reduction and an ablation therapy visualization framework for CT-guided ablation therapy of liver cancer. The experiment results indicate that our approach can reduce metal artifacts and improve image quality. Furthermore, our proposed method demonstrates the potential for displaying the relative position of the tumor and the needle intraoperatively.









Similar content being viewed by others
References
Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, Laversanne M, McGlynn KA, Soerjomataram I (2022) Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol 77:1598–1606. https://doi.org/10.1016/j.jhep.2022.08.021
Gillams AR (2004) Liver ablation therapy. Br J Radiol 77:713–723. https://doi.org/10.1259/bjr/86761907
Meloni MF, Chiang J, Laeseke PF, Dietrich CF, Sannino A, Solbiati M, Nocerino E, Brace CL, Lee FT (2017) Microwave ablation in primary and secondary liver tumours: technical and clinical approaches. Int J Hyperth 33:15–24. https://doi.org/10.1080/02656736.2016.1209694
Wellenberg RHH, Hakvoort ET, Slump CH, Boomsma MF, Maas M, Streekstra GJ (2018) Metal artifact reduction techniques in musculoskeletal CT-imaging. Eur J Radiol 107:60–69. https://doi.org/10.1016/j.ejrad.2018.08.010
Gjesteby L, Yang Q, Xi Y, Shan H, Claus B, Jin Y, De Man B, Wang G (2017) Deep learning methods for CT image-domain metal artifact reduction. Developments in X-ray Tomography XI. pp 147–152
Gjesteby L, Shan H, Yang Q, Xi Y, Jin Y, Giantsoudi D, Paganetti H, De Man B, Wang G (2019) A dual-stream deep convolutional network for reducing metal streak artifacts in CT images. Phys Med Biol 64:235003
Gjesteby L, Yang Q, Xi Y, Zhou Y, Zhang J, Wang G (2017) Deep learning methods to guide CT image reconstruction and reduce metal artifacts. Medical Imaging 2017: Physics of Medical Imaging. pp 752–758
Xia H, Jian W, Fan T, Tao Z, Yu Z (2018) Metal artifact reduction on cervical CT images by deep residual learning. Biomed Eng Online 17:1–15. https://doi.org/10.1186/s12938-018-0609-y
Zhu LL, Han Y, Li L, Xi XQ, Zhu MW, Yan B (2019) Metal artifact reduction for X-Ray computed tomography using U-Net in image domain. Ieee Access 7:98743–98754. https://doi.org/10.1109/access.2019.2930302
Wang J, Noble JH, Dawant BM (2019) Metal artifact reduction for the segmentation of the intra cochlear anatomy in CT images of the ear with 3D-conditional GANs. Med Image Anal 58: 101553. https://doi.org/10.1016/j.media.2019.101553
Gjesteby LA, Shan H, Yang Q, Xi Y, Claus BEH, Jin Y, Man BD, Wang G (2018) Deep neural network for CT metal artifact reduction with a perceptual loss function.
Liao H, Lin W-A, Yuan J, Zhou SK, Luo J (2020) ADN: artifact disentanglement network for unsupervised metal artifact reduction. IEEE Trans Med Imaging 39:634–643
Lin W-A, Liao H, Peng C, Sun X, Zhang J, Luo J, Chellappa R, Zhou SK (2019) DuDoNet: dual domain network for CT metal artifact reduction. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp 10504–10513
Lee J, Gu J, Ye JC (2021) Unsupervised CT metal artifact learning using attention-guided β-CycleGAN. IEEE Trans Med Imaging 40:3932–3944. https://doi.org/10.1109/tmi.2021.3101363
Huang Z, Zhang G, Lin J, Pang Y, Wang H, Bai T, Zhong L (2022) Multi-modal feature-fusion for CT metal artifact reduction using edge-enhanced generative adversarial networks. Comput Methods Programs Biomed 217:106700
Wang Z, Vandersteen C, Demarcy T, Gnansia D, Raffaelli C, Guevara N, Delingette H (2021) Inner-ear augmented metal artifact reduction with simulation-based 3D generative adversarial networks. Comp Med Imaging Graphics Official J Computerized Med Imaging Soc 93:101990
Lyu Y, Lin W-A, Liao H, Lu J, Zhou SK (2020) Encoding metal mask projection for metal artifact reduction in computed tomography. MICCAI 2020:147–157
Sun B, Jia S, Jiang X, Jia F (2023) Double U-Net CycleGAN for 3D MR to CT image synthesis. Int J Comput Assist Radiol Surg 18:149–156. https://doi.org/10.1007/s11548-022-02732-x
Qiu W, Yuchi M, Ding M, Tessier D, Fenster A (2013) Needle segmentation using 3D Hough transform in 3D TRUS guided prostate transperineal therapy. Med Phys 40: 042902. https://doi.org/10.1118/1.4795337
Chen L-C, Zhu Y, Papandreou G, Schroff F, Adam H (2018) Encoder–decoder with atrous separable convolution for sSemantic image segmentation. In: Computer Vision—ECCV 2018, pp 833–851
Ledig C, Theis L, Huszár F, Caballero J, Cunningham A, Acosta A, Aitken A, Tejani A, Totz J, Wang Z, Shi W (2017) Photo-realistic single image super-resolution using a generative adversarial network. In: 2017 IEEE Conference on computer vision and pattern recognition (CVPR), pp 105–114
Isola P, Zhu JY, Zhou T, Efros AA (2017) Image-to-image translation with conditional adversarial networks. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR), pp 5967–5976
Johnson J, Alahi A, Li FF (2016) Perceptual losses for real-time style transfer and super-resolution. In: 14th European conference on computer vision (ECCV), pp 694–711
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: International conference on learning representations (ICLR).
Lee TC, Kashyap RL, Chu CN (1994) Building skeleton models via 3-D medial surface axis thinning algorithms. CVGIP: Graphical Models and Image Processing 56: 462–478. https://doi.org/10.1006/cgip.1994.1042
Mukhopadhyay P, Chaudhuri BB (2015) A survey of Hough transform. Pattern Recogn 48:993–1010. https://doi.org/10.1016/j.patcog.2014.08.027
Barva M, Uhercik M, Mari JM, Kybic J, Duhamel JR, Liebgott H, Hlavac V, Cachard C (2008) Parallel integral projection transform for straight electrode localization in 3-D ultrasound images. IEEE Trans Ultrason Ferroelectr Freq Control 55:1559–1569. https://doi.org/10.1109/TUFFC.2008.833
Larsen ABL, Sønderby SK, Larochelle H, Winther O (2016) Autoencoding beyond pixels using a learned similarity metric. In: 33rd International Conference on Machine Learning(PMLR), pp 1558--1566
Zhao H, Gallo O, Frosio I, Kautz J (2017) Loss functions for image restoration with neural networks. IEEE Trans Comput Imaging 3:47–57. https://doi.org/10.1109/TCI.2016.2644865
Arabi H, Zaidi H (2021) Deep learning–based metal artefact reduction in PET/CT imaging. Eur Radiol 31:6384–6396
Selles M, Slotman DJ, van Osch JA, Nijholt IM, Wellenberg RH, Maas M, Boomsma MF (2023) Is AI the way forward for reducing metal artifacts in CT? Development of a generic deep learning-based method and initial evaluation in patients with sacroiliac joint implants. Eur J Radiol 163:110844
Izzo F, Granata V, Grassi R, Fusco R, Palaia R, Delrio P, Carrafiello G, Azoulay D, Petrillo A, Curley SA (2019) Radiofrequency ablation and microwave ablation in liver tumors: an update. Oncologist 24:E990–E1005. https://doi.org/10.1634/theoncologist.2018-0337
Mehrtash A, Ghafoorian M, Pernelle G, Ziaei A, Heslinga FG, Tuncali K, Fedorov A, Kikinis R, Tempany CM, Wells WM, Abolmaesumi P, Kapur T (2019) Automatic needle segmentation and localization in MRI with 3-D convolutional neural networks: application to MRI-targeted prostate biopsy. IEEE Trans Med Imaging 38:1026–1036. https://doi.org/10.1109/tmi.2018.2876796
Zhao S, Dong Y, Chang E, Xu Y (2019) Recursive cascaded networks for unsupervised medical image registration. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV), pp 10599–10609
Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV (2019) VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging 38:1788–1800. https://doi.org/10.1109/TMI.2019.2897538
Acknowledgements
This work was supported in part by the Ministry of Science and Technology of the People’s Republic of China under Grant 2016YFC0106201, in part by the Shanghai Science and Technology Commission of Shanghai Municipality under Grant 19DZ2280300, in part by Shanghai Jiao Tong University Medical Engineering Cross Research Funds under Grant YG2021ZD05, in part by Shanghai Hospital Development Center Foundation under Grant SHDC12021112, in part by Shanghai Zhangjiang National Independent Innovation Demonstration Zone Special Development Fund Major Project under Grant ZJ2021-ZD-007.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the Ethics Committee of Fudan University Shanghai Cancer Center.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liang, D., Zhang, S., Zhao, Z. et al. Two-stage generative adversarial networks for metal artifact reduction and visualization in ablation therapy of liver tumors. Int J CARS 18, 1991–2000 (2023). https://doi.org/10.1007/s11548-023-02986-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-023-02986-z