Abstract
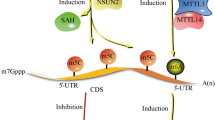
RNA 5-methylcytosine (m5C) sites perform a major role in numerous biological processes and commonly reported in both DNA and RNA cellular. The enzymatic mechanism and biological functions of m5C sites in DNA remain the focusing area of researchers for last few decades. Likewise, the investigators also targeted m5C sites in RNA due to its cellular functions, positioning and formation mechanism. Currently, several rudimentary roles of the m5C in RNA have been explored, but a lot of improvements are still under consideration. Initially, the identification of RNA methylcytosine sites was carried out via experimental methods, which were very hard, erroneous and time consuming owing to partial availability of recognized structures. Looking at the significance of m5C role in RNA, scientists have diverted their attention from structure to sequence-based prediction. In this regards, an intelligent computational model is proposed in order to identify m5C sites in RNA with high precision. Three RNA sequences formulation methods namely: pseudo dinucleotide composition,pseudo trinucleotide composition and pseudo tetra nucleotide composition are applied to extract variant and high profound numerical features. In a sequel, the vector spaces are fused to build a hybrid space in order to compensate the weakness of each other. Various learning hypotheses are examined to select the best operational engine, which can truly identify the pattern of the target class. The strength and generalization of the proposed model are measured using two different cross validation tests. The reported outcomes reveal that the proposed model achieved 3% better accuracy than that of the highest present approach in the literature so far.
Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Yue Y, Liu J, He C. RNA N6-mefhyladenosine methylation in post-transcriptional gene expression regulation. Genes & Development, 2015, 29(29): 1343–1355
Edelheit S, Schwartz S, Mumbach M R, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m C within archaeal mRNAs. PLoS Genetics, 2013, 9(9): el003602
Feng P, Ding H, Chen W, Lin H. Identifying RNA 5-mefhylcytosine sites via pseudo nucleotide compositions. Molecular BioSystems, 2016, 12(12): 3307–3311
Agris P F. Bringing order to translation: the contributions of trans fer RNA anticodon-domain modifications. EMBO Reports, 2008, 9(9): 629–635
Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Research, 2006, 34(34): 721–733
Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Bio chemistry, 2010, 49(49): 4934 1944
Schaefer M, Pollex T, Hanna K, Lyko F RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Research, 2008, 37(37): e12
Hussain S, Sajini A A, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson J G, Odom D T, Ule J. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Reports, 2013, 4(4): 255–261
Zou Q, Guo J, Ju Y, Wu M, Zeng X, Hong Z. Improving tRNAscan-SE annotation results via ensemble classifiers. Molecular Informatics, 2015, 34(11-12): 761–770
Khoddami V, Cairns B R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nature Biotechnology, 2013, 31(31): 458 164
Feng P, Ding H, Yang H, Chen W, Lin H, Chou K-C. iRNA-PseColl: identifying the occurrence sites of different RNA modifications by in corporating collective effects of nucleotides into PseKNC Molecular Therapy-Nucleic Acids, 2017, 7: 155–163
Wan S, Duan Y, Zou Q. HPSLPred: an ensemble multi-label classifier for human protein subcellular location prediction with imbalanced source. Proteomics, 2017, 17(17-18): 1700262
Liao Z, Ju Y, Zou Q. Prediction of G protein-coupled receptors with SVM-prot features and random forest. Scientifica, 2016, 2016: 8309253
Chen W, Xing P, Zou Q. Detecting N 6-mefhyladenosine sites from RNA transcriptomes using ensemble support vector machines. Scien tific Reports, 2017, 7: 40242
Lin C, Zou Y, Qin J, Liu X, Jiang Y, Ke C, Zou Q. Hierarchical classification of protein folds using a novel ensemble classifier. PLoS One, 2013, 8(8): e56499
Zhang M, Y, Li L, Liu Z, Yang X, Yu D J. Accurate RNA 5-methylcytosine site prediction based on heuristic physical-chemical properties reduction and classifier ensemble. Analytical Biochemistry, 2018, 550: 41–48
Qiu W R, Jiang S Y, Xu Z C, Xiao X, Chou K C. iRNAm5C-PseDNC identifying RNA 5-mefhylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget, 2017, 8(25): 41178
Iqbal M, Hayat M. “iSS-Hyb-mRMR”: identification of splicing sites using hybrid space of pseudo trinucleotide and pseudo tetranucleotide composition. Computer Methods and Programs in Biomedicine, 2016, 128: 1–11
Squires J E, Patel H R, Nousch M, Sibbritt T, Humphreys D T, Parker B J, Suter C M, Preiss T. Widespread occurrence of 5-mefhylcytosine in human coding and non-coding RNA. Nucleic Acids Research, 2012, 40(40): 5023–5033
Sun W J, Li J H, Liu S, Wu J, Zhou H, Qu L H, Yang J H RMBase: a resource for decoding the landscape of RNA modifications from high- throughput sequencing data. Nucleic Acids Research, 2015, 44(D1): D259–D265
Fu L, Niu B, Zhu Z, Wu S, Li W CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 2012, 28(28): 3150–3152
Akbar S, Hayat M, Iqbal M, Jan M A. iACP-GAEnsC: evolutionary genetic algorithm based ensemble classification of anticancer peptides by utilizing hybrid feature space. Artificial Intelligence in Medicine, 2017, 79: 62–70
Hayat M, Khan A. Predicting membrane protein types by fusing com posite protein sequence features into pseudo amino acid composition. Journal of Theoretical Biology, 2011, 271(271): 10–17
Kabir M, Yu D J. Predicting DNase I hypersensitive sites via un-biased pseudo trinucleotide composition. Chemometrics and Intelligent Lab oratory Systems, 2017, 167: 78–84
Tahir M, Hayat M, Kabir M. Sequence based predictor for discrim ination of enhancer and their types by applying general form of Chou's trinucleotide composition. Computer Methods and Programs in Biomedicine, 2017, 146: 69–75
Liu Z, Xiao X, Qiu W R, Chou K C. iDNA-methyl: identifying DNA methylation sites via pseudo trinucleotide composition. Analytical Bio chemistry, 2015, 474: 69–77
Kabir M, Hayat M. iRSpot-GAEnsC: identifing recombination spots via ensemble classifier and extending the concept of Chou's PseAAC to formulate DNA samples. Molecular Genetics and Genomics, 2016, 291(291): 285–296
Chen W, Lei T Y, Jin D C, Lin H, Chou K C. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Analyti cal Biochemistry, 2014, 456: 53–60
Hayat M, Khan A. WRF-TMH: predicting transmembrane helix by fus ing composition index and physicochemical properties of amino acids. Amino Acids, 2013, 44(44): 1317–1328
Ali F, Hayat M. Classification of membrane protein types using voting feature interval in combination with Chou's pseudo amino acid com position. Journal of Theoretical Biology, 2015, 384: 78–83
Akbar S, Hayat M. iMethyl-STTNC: identification of N6- methyladenosine sites by extending the idea of SAAC into Chou's PseAAC to formulate RNA sequences. Journal of Theoretical Biology, 2018, 455: 205–211
Khan A, Majid A, Hayat M. CE-PLoc: an ensemble classifier for predicting protein subcellular locations by fusing different modes of pseudo amino acid composition. Computational Biology and Chem istry, 2011, 35(35): 218–229
Hu J, Han K, Li Y, Yang J Y, Shen H B, Yu D J. TargetCrys: pro tein crystallization prediction by fusing multi-view features with two- layered SVM. Amino Acids, 2016, 48(48): 2533–2547
Hayat M, Khan A. Discriminating outer membrane proteins with fuzzy K-nearest neighbor algorithms based on the general form of Chou's PseAAC Protein and Peptide Letters, 2012, 19(19): 411–421
Ahmad S, Kabir M, Hayat M. Identification of heat shock protein families and J-protein types by incorporating dipeptide composition into Chou's general PseAAC. Computer Methods and Programs in Biomedicine, 2015, 122(122): 165–174
Liu B, Wang S, Long R, Chou K C. iRSpot-EL: identify recombina tion spots with an ensemble learning approach. Bioinformatics, 2016, 33(33): 35–41
Xiao X, Min J L, Lin W Z, Liu Z, Cheng X, Chou K C. iDrug- target: predicting the interactions between drug compounds and tar get proteins in cellular networking via benchmark dataset optimiza tion approach. Journal of Biomolecular Structure and Dynamics, 2015, 33(33): 2221–2233
Akbar S, Hayat M, Kabir M, Iqbal M. iAFP-gap-SMOTE: an efficient feature extraction scheme gapped dipeptide composition is coupled with an oversampling technique for identification of antifreeze pro teins. Letters in Organic Chemistry, 2019, 16(16): 294–302
Lin W Z, Fang J A, Xiao X, Chou K C. iDNA-Prot: identification of DNA binding proteins using random forest with grey model. PLoS One, 2011, 6(9): e24756
Huang Y F, Chiu L Y, Huang C C, Huang C K. Predicting RNA- binding residues from evolutionary information and sequence conser vation. BMC Genomics, 2010, 11(11): S2
Chen W, Ding H, Feng P, Lin H, Chou K C. iACP: a sequence- based tool for identifying anticancer peptides. Oncotarget, 2016, 7(7): 16895
Akbar S, Ahmad A, Hayat M, Ah F Face recognition using hybrid feature space in conjunction with support vector machine. Journal of Applied Environmental and Biological Sciences, 2015, 5(5): 28–36
Hu J, Yan X. BS-KNN: an effective algorithm for predicting protein subchloroplast localization. Evolutionary Bioinformatics Online, 2012, 8: 79
Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Statistics Surveys, 2010, 4: 40–79
Ng A Y. Preventing “overfitting” of cross-validation data. In: Proceed ings of the 14th International Conference on Machine Learning. 1997, 245–253
Vehtari A, Gelman A, Gabry J. Practical Bayesian model evaluation using leave-one-out cross-validation and WAIC Statistics and Com puting, 2017, 27(27): 1413–1432
Ahmad J, Javed F, Hayat M. Intelligent computational model for clas sification of sub-Golgi protein using oversampling and fisher feature selection methods. Artificial Intelligence in Medicine, 2017, 78: 14–22
Tahir M, Hayat M. Machine learning based identification of protein- protein interactions using derived features of physiochemical properties and evolutionary profiles. Artificial Intelligence in Medicine, 2017, 78: 61–71
Zhang W, Robbins K, Wang Y, Bertrand K, Rekaya R. A jackknife-like method for classification and uncertainty assessment of multi-category tumor samples using gene expression information. BMC Genomics, 2010, 11(11): 273
Elloumi M, Iliopoulos C, Wang J T, Zomaya A Y. Pattern Recognition in Computational Molecular Biology: Techniques and Approaches. John Wiley & Sons, 2015
Wasserman L. All of Statistics: a Concise course in Statistical Infer ence. Springer Science & Business Media, 2013
Bengio Y, Grandvalet Y. No unbiased estimator of the variance of K- fold cross-validation. Journal of Machine Learning Research, 2004, 5(Sep): 1089–1105
Kohavi R. A study of cross-validation and bootstrap for accuracy esti mation and model selection. In: Proceedings of the 14th International Joint Conference on Artificial Intellgence-Volum 2. 1995, 1137–1145
Fushiki T. Estimation of prediction error by using K-fold cross- validation. Statistics and Computing, 2011, 21(21): 137–146
Doreswamy H K. Performance evaluation of predictive classifiers for knowledge discovery from engineering materials data sets. 2012, arXiv preprint arXiv: 1209.2501
Qiu W R, Xiao X, Lin W Z, Chou K C. iMethyl-PseAAC: identifica tion of protein methylation sites via a pseudo amino acid composition approach. BioMed Research International, 2014, 2014: 947416
Xiao X, Wang P, Chou K C. iNR-PhysChem: a sequence-based predic tor for identifying nuclear receptors and their subfamilies via physical- chemical property matrix. PLoS One, 2012, 7(7): e30869
Xiao X, Wang P, Lin W Z, Jia J H, Chou K C. iAMP-2L: a two-level multi-label classifier for identifying antimicrobial peptides and their functional types. Analytical Biochemistry, 2013, 436(436): 168–177
Feng P, Yang H, Ding H, Lin H, Chen W, Chou K C. iDNA6mA- PseKNC: identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC Genomics, 2019, 111(111): 96–102
Chen W, Yang H, Feng P, Ding H, Lin H. iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical proper ties. Bioinformatics, 2017, 33(33): 3518–3523
Zhao Y W, Su Z D, Yang W, Lin H, Chen W, Tang H. IonchanPred 2.0: a tool to predict Ion channels and their types. International Journal of Molecular Sciences, 2017, 18(18): 1838
Dao F Y, Yang H, Su Z D, Yang W, Wu Y, Hui D, Chen W, Tang H, Lin H. Recent advances in conotoxin classification by using machine learning methods. Molecules, 2017, 22(22): 1057
Acknowledgments
We thank to the anonymous reviewers for their careful reading of our manuscript and their useful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Shahid Akbar received his Bachelor degree in Computer Science & Information Technology from Islamic University of Technology, Bangladesh in 2011. He received his MS degree in Computer Science from Abdul Wali Khan University (AWKU), Pakistan in 2015. He is currently pursuing his PhD in Computer Science from Abdul Wali Khan University (AWKU), Pakistan. His research interests include bioinformatics, pattern recognition and machine learning.
Maqsood Hayat received his MCS degree from Gomal University, Pakistan in 2004 and his MS degree in Software & System Engineering from Mohammad Ali Jinnah University (MAJU), Pakistan in 2009. He received his PhD degree from Department of Computer & Information Sciences, Pakistan Institute of Engineering & Applied Sciences, Pakistan. He is working as an assistant professor since august 2012. His main research includes machine learning, pattern recognition, evolutionary computing and its application in bioinformatics.
Muhammad Iqbal received his Bachelor degree in Computer Science from Islamia College University, Pakistan in 2012. He is currently pursuing his MS degree in Computer Science from Abdul Wali Khan University (AWKUM), Pakistan. His research interests include bioinformatics, pattern recognition and machine learning.
Muhammad Tahir received his PhD degree in Computer Science from Abdul Wali Khan University (AWKUM), Pakistan in 2017. He is working as a lecturer since August 2010. His main research includes pattern recognition, bioinformatics and machine learning.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Akbar, S., Hayat, M., Iqbal, M. et al. iRNA-PseTNC: identification of RNA 5-methylcytosine sites using hybrid vector space of pseudo nucleotide composition. Front. Comput. Sci. 14, 451–460 (2020). https://doi.org/10.1007/s11704-018-8094-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11704-018-8094-9