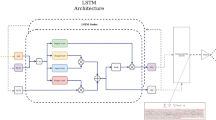
Abstract
A promoter is a short region of DNA that can bind RNA polymerase and initiate gene transcription. It is usually located directly upstream of the transcription initiation site. DNA promoters have been proven to be the main cause of many human diseases, especially diabetes, cancer or Huntington’s disease. Therefore, the classification of promoters has become an interesting problem and has attracted the attention of many researchers in the field of bioinformatics. Various studies have been conducted in order to solve this problem, but their performance still needs further improvement. In this research, we segmented the DNA sequence in a k-mers manner, then trained the word vector model, inputted it into long short-term memory(LSTM) and used the attention mechanism to predict. Our method can achieve 93.45% and 90.59% cross-validation accuracy in the two layers, respectively. Our results are better than others based on the same data set, and provided some ideas for accurately predicting promoters. In addition, this research suggested that natural language processing can play a significant role in biological sequence prediction.
Similar content being viewed by others
References
Liu B, Li K. iPromoter-2L2.0: identifying promoters and their types by combining smoothing cutting window algorithm and sequence-based features. Molecular Therapy Nucleic Acids, 2019, 18: 80–87
He W, Jia C, Duan Y, Zou Q. 70ProPred: a predictor for discovering sigma70 promoters based on combining multiple features. BMC Systems Biology, 2018, 12(4): 44
Xu Y, Zhao W, Olson S D, Prabhakara K S, Zhou X. Alternative splicing links histone modifications to stem cell fate decision. Genome Biology, 2018, 19(1): 133
Xu Y, Wang Y, Luo J, Zhao W, Zhou X. Deep learning of the splicing (epi) genetic code reveals a novel candidate mechanism linking histone modifications to ESC fate decision. Nucleic Acids Research, 2017, 45(21): 12100–12112
Zhao Y, Wang F, Juan L. MicroRNA promoter identification in Arabidopsis using multiple histone markers. BioMed Research International, 2015, 2015: 861402
Zhao Y, Wang F, Chen S, Wan J, Wang G. Methods of MicroRNA promoter prediction and transcription factor mediated regulatory network. BioMed Research International, 2017, 2017: 7049406
Wang G, Wang Y, Teng M, Zhang D, Li L, Liu Y. Signal transducers and activators of transcription-1 (STAT1) regulates microRNA transcription in interferon γ-stimulated HeLa cells. PLoS One, 2010, 5(7): e11794
Liu B, Han L, Liu X, Wu J, Ma Q. Computational prediction of sigma-54 promoters in bacterial genomes by integrating motif finding and machine learning strategies. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2019, 16(4): 1211–1218
Chen J, Zhang S. Integrative cancer genomics: models, algorithms and analysis. Frontiers of Computer Science, 2017, 11(3): 392–406
Sun J, Du P F. Predicting protein subchloroplast locations: the 10th anniversary. Frontiers of Computer Science, 2021, 15(2): 152901
Li Q Z, Lin H. The recognition and prediction of σ70 promoters in Escherichia coli K-12. Journal of Theoretical Biology, 2006, 242(1): 135–141
Song K. Recognition of prokaryotic promoters based on a novel variable-window Z-curve method. Nucleic Acids Research, 2012, 40(3): 963–971
de Avila e Silva S, Forte F, Sartor I T S, Andrighetti T, Gerhardt G J L, Delamare A P L, Echeverrigaray S. DNA duplex stability as discriminative characteristic for Escherichia coli σ54- and σ28-dependent promoter sequences. Biologicals, 2014, 42(1): 22–28
Lin H, Deng E Z, Ding H, Chen W, Chou K C. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Research, 2014, 42(21): 12961–12972
Liu B, Yang F, Huang D S, Chou K C. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics, 2018, 34(1): 33–40
Xiao X, Xu Z C, Qiu W R, Wang P, Ge H T, Chou K C. iPSW(2L)-PseKNC: a two-layer predictor for identifying promoters and their strength by hybrid features via pseudo K-tuple nucleotide composition. Genomics, 2019, 111(6): 1785–1793
Le N Q K, Yapp E K Y, Nagasundaram N, Yeh H Y. Classifying promoters by interpreting the hidden information of DNA sequences via deep learning and combination of continuous FastText N-grams. Frontiers in Bioengineering and Biotechnology, 2019, 7: 705
Zhang Z Y, Yang Y H, Ding H, Wang D, Chen W, Lin H. Design powerful predictor for mRNA subcellular location prediction in Homo sapiens. Briefings in Bioinformatics, 2021, 22(1): 526–535
Lin H, Liang Z Y, Tang H, Chen W. Identifying sigma70 promoters with novel pseudo nucleotide composition. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2019, 16(4): 1316–1321
Lai H Y, Zhang Z Y, Su Z D, Su W, Ding H, Chen W, Lin H. iProEP: a computational predictor for predicting promoter. Molecular Therapy Nucleic Acids, 2019, 17: 337–346
Wang J, Chen S, Dong L, Wang G. CHTKC: a robust and efficient k-mer counting algorithm based on a lock-free chaining hash table. Briefings in Bioinformatics, 2021, 22(3): bbaa063
Liu B, Gao X, Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Research, 2019, 47(20): e127
Mikolov T, Sutskever I, Chen K, Corrado G, Dean J. Distributed representations of words and phrases and their compositionality. In: Proceedings of the 26th International Conference on Neural Information Processing Systems. 2013, 3111–3119
Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. 2013, arXiv preprint arXiv: 1301.3781
Zou Q, Xing P, Wei L, Liu B. Gene2vec: gene subsequence embedding for prediction of mammalian N6 — methyladenosine sites from mRNA. RNA, 2019, 25(2): 205–218
Chen J, Zou Q, Li J. DeepM6ASeq-EL: prediction of human N6-methyladenosine (m6A) sites with LSTM and ensemble learning. Frontiers of Computer Science, 2022, 16(2): 162302
Zhao X, Jiao Q, Li H, Wu Y, Wang H, Huang S, Wang G. ECFS-DEA: an ensemble classifier-based feature selection for differential expression analysis on expression profiles. BMC Bioinformatics, 2020, 21(1): 43
Tang Y J, Pang Y H, Liu B. IDP-Seq2Seq: identification of intrinsically disordered regions based on sequence to sequence learning. Bioinformatics, 2020, 36(21): 5177–5186
Du Y, Chen Z, Zhang C, Cao X. Research on axial bearing capacity of rectangular concrete-filled steel tubular columns based on artificial neural networks. Frontiers of Computer Science, 2017, 11(5): 863–873
Hayward S. Risk aversion and agents’ survivability in a financial market. Frontiers of Computer Science in China, 2009, 3(2): 158–166
Wang Z, He W, Tang J, Guo F. Identification of highest-affinity binding sites of yeast transcription factor families. Journal of Chemical Information and Modeling, 2020, 60(3): 1876–1883
Wang H, Ding Y, Tang J, Guo F. Identification of membrane protein types via multivariate information fusion with Hilbert-Schmidt Independence Criterion. Neurocomputing, 2020, 383: 257–269
Li J, Pu Y, Tang J, Zou Q, Guo F. DeepAVP: a dual-channel deep neural network for identifying variable-length antiviral peptides. IEEE Journal of Biomedical and Health Informatics, 2020, 24(10): 3012–3019
Shen Y, Tang J, Guo F. Identification of protein subcellular localization via integrating evolutionary and physicochemical information into Chou’s general PseAAC. Journal of Theoretical Biology, 2019, 462: 230–239
Su R, Wu H, Xu B, Liu X, Wei L. Developing a multi-dose computational model for drug-induced hepatotoxicity prediction based on toxicogenomics data. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2019, 16(4): 1231–1239
Wei L, Chen H, Su R. M6APred-EL: a sequence-based predictor for identifying N6-methyladenosine sites using ensemble learning. Molecular Therapy Nucleic Acids, 2018, 12: 635–644
Wei L, Wan S, Guo J, Wong K K L. A novel hierarchical selective ensemble classifier with bioinformatics application. Artificial Intelligence in Medicine, 2017, 83: 82–90
Wei L, Xing P, Zeng J, Chen J, Su R, Guo F. Improved prediction of protein—protein interactions using novel negative samples, features, and an ensemble classifier. Artificial Intelligence in Medicine, 2017, 83: 67–74
Xu L, Liang G, Chen B, Tan X, Xiang H, Liao C. A computational method for the identification of endolysins and autolysins. Protein & Peptide Letters, 2020, 27(4): 329–336
Xu L, Liang G, Liao C, Chen G D, Chang C C. An efficient classifier for alzheimer’s disease genes identification. Molecules, 2018, 23(12): 3140
Xu L, Liang G, Liao C, Chen G D, Chang C C. k-Skip-n-Gram-RF: a random forest based method for alzheimer’s disease protein identification. Frontiers in Genetics, 2019, 10: 33
Chen W, Feng P, Song X, Lv H, Lin H. iRNA-m7G: identifying N7-methylguanosine sites by fusing multiple features. Molecular therapy Nucleic Acids, 2019, 18: 269–274
Liu K, Chen W. iMRM: a platform for simultaneously identifying multiple kinds of RNA modifications. Bioinformatics, 2020, 36(11): 3336–3342
Wang G, Wang Y, Feng W, Wang X, Yang J Y, Zhao Y, Wang Y, Liu Y. Transcription factor and microRNA regulation in androgen-dependent and -independent prostate cancer cells. BMC Genomics, 2008, 9(S2): S22
Wang G, Luo X, Wang J, Wan J, Xia S, Zhu H, Qian J, Wang Y. MeDReaders: a database for transcription factors that bind to methylated DNA. Nucleic Acids Research, 2018, 46(D1): D146–D151
Liu B, Luo Z, He J. sgRNA-PSM: predict sgRNAs on-target activity based on position-specific mismatch. Molecular Therapy Nucleic Acids, 2020, 20: 323–330
Jiao Y, Du P. Performance measures in evaluating machine learning based bioinformatics predictors for classifications. Quantitative Biology, 2016, 4(4): 320–330
Li Q, XU L, Li Q, Zhang L. Identification and classification of enhancers using dimension reduction technique and recurrent neural network. Computational and Mathematical Methods in Medicine, 2020, 2020: 8852258
Li Q, Dong B, Wang D, Wang S. Identification of secreted proteins from malaria protozoa with few features. IEEE Access, 2020, 8: 89793–89801
Li Q, Zhou W, Wang D, Wang S, Li Q. Prediction of anticancer peptides using a low-dimensional feature model. Frontiers in Bioengineering and Biotechnology, 2020, 8: 892
Meng C, Guo F, Zou Q. CWLy-SVM: a support vector machine-based tool for identifying cell wall lytic enzymes. Computational Biology and Chemistry, 2020, 87: 107304
Wang Y, Shi F, Cao L, Dey N, Wu Q, Ashour A S, Sherratt R S, Rajinikanth V, Wu L. Morphological segmentation analysis and texture-based support vector machines classification on mice liver fibrosis microscopic images. Current Bioinformatics, 2019, 14(4): 282–294
Meng C, Jin S, Wang L, Guo F, Zou Q. AOPs-SVM: a sequence-based classifier of antioxidant proteins using a support vector machine. Frontiers in Bioengineering and Biotechnology, 2019, 7: 224
Zhang N, Sa Y, Guo Y, Lin W, Wang P, Feng Y. Discriminating ramos and jurkat cells with image textures from diffraction imaging flow cytometry based on a support vector machine. Current Bioinformatics, 2018, 13(1): 50–56
Shen Y, Ding Y, Tang J, Zou Q, Guo F. Critical evaluation of web-based prediction tools for human protein subcellular localization. Briefings in Bioinformatics, 2020, 21(5): 1628–1640
Shen C, Ding Y, Tang J, Jiang L, Guo F. LPI-KTASLP: prediction of LncRNA-protein interaction by semi-supervised link learning with multivariate information. IEEE Access, 2019, 7: 13486–13496
Ding Y, Tang J, Guo F. Identification of drug-side effect association via semisupervised model and multiple kernel learning. IEEE Journal of Biomedical and Health Informatics, 2019, 23(6): 2619–2632
Ding Y, Tang J, Guo F. Identification of drug-side effect association via multiple information integration with centered kernel alignment. Neurocomputing, 2019, 325: 211–224
Qiang X, Zhou C, Ye X, Du P F, Su R, Wei L. CPPred-FL: a sequence-based predictor for large-scale identification of cell-penetrating peptides by feature representation learning. Briefings in Bioinformatics, 2020, 21(1): 11–23
Wei L, Zhou C, Chen H, Song J, Su R. ACPred-FL: a sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics, 2018, 34(23): 4007–4016
Xu L, Liang G, Shi S, Liao C. SeqSVM: a sequence-based support vector machine method for identifying antioxidant proteins. International Journal of Molecular Sciences, 2018, 19(6): 1773
Xu L, Liang G, Wang L, Liao C. A novel hybrid sequence-based model for identifying anticancer peptides. Genes, 2018, 9(3): 158
Jiang Q, Wang G, Jin S, Li Y, Wang Y. Predicting human microRNA-disease associations based on support vector machine. International Journal of Data Mining and Bioinformatics, 2013, 8(3): 282–293
Wang Y, Liu K, Ma Q, Tan Y, Du W, Lv Y, Tian Y, Wang H. Pancreatic cancer biomarker detection by two support vector strategies for recursive feature elimination. Biomarkers in Medicine, 2019, 13(2): 105–121
Huo Y, Xin L, Kang C, Wang M, Ma Q, Yu B. SGL-SVM: a novel method for tumor classification via support vector machine with sparse group Lasso. Journal of Theoretical Biology, 2020, 486: 110098
Liu B, Li C C, Yan K. DeepSVM-fold: protein fold recognition by combining Support Vector machines and pairwise sequence similarity scores generated by deep learning networks. Briefings in Bioinformatics, 2020, 21(5): 1733–1741
Li C C, Liu B. MotifCNN-fold: protein fold recognition based on fold-specific features extracted by motif-based convolutional neural networks. Briefings in Bioinformatics, 2020, 21(6): 2133–2141
Acknowledgements
The classifiers in this article were provided by the WEKA platform. This research was funded by the Natural Science Foundation of China (Grant No. 61902259), the Natural Science Foundation of Guangdong province (2018A0303130084). The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Qingwen Li is a doctoral student at the Institute of Biophysics, Chinese Academy of Sciences, China. He once visited the University of Electronic Science and Technology of China, China. His research interests include bioinformatics and neurobiology.
Lichao Zhang is a lecturer at the School of Intelligent Manufacturing and Equipment, Shenzhen Institute of Information Technology, China. Her research interests include machine learning and bioinformatics.
Lei Xu is an associate professor at the School of Electronic and Communication Engineering, Shenzhen Polytechnic, China. Her research interests include machine learning and bioinformatics.
Quan Zou is a professor at the University of Electronic Science and Technology of China, China. He is a senior member of IEEE and ACM. He won the Clarivate Analytics Highly Cited Researchers in 2018 and 2019. He majors in bioinformatics, machine learning, and algorithms.
Jin Wu is a lecture at the School of Management, Shenzhen Polytechnic. China. Her research interests include microbiome and bioinformatics.
Qingyuan Li is a senior engineer of Forestry and Fruit Tree Research Institute, Wuhan Academy of Agricultural Sciences, China. His research interests include plant genetics, plant genomics and bioinformatics.
Electronic Supplementary Material
Rights and permissions
About this article
Cite this article
Li, Q., Zhang, L., Xu, L. et al. Identification and classification of promoters using the attention mechanism based on long short-term memory. Front. Comput. Sci. 16, 164348 (2022). https://doi.org/10.1007/s11704-021-0548-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11704-021-0548-9