Abstract
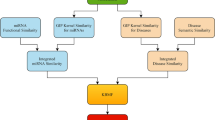
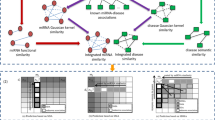
Numerous studies have demonstrated that human microRNAs (miRNAs) and diseases are associated and studies on the microRNA-disease association (MDA) have been conducted. We developed a model using a low-rank approximation-based link propagation algorithm with Hilbert–Schmidt independence criterion-based multiple kernel learning (HSIC-MKL) to solve the problem of the large time commitment and cost of traditional biological experiments involving miRNAs and diseases, and improve the model effect. We constructed three kernels in miRNA and disease space and conducted kernel fusion using HSIC-MKL. Link propagation uses matrix factorization and matrix approximation to effectively reduce computation and time costs. The results of the experiment show that the approach we proposed has a good effect, and, in some respects, exceeds what existing models can do.
Similar content being viewed by others
References
Shi H, Zhang G, Zhou M, Cheng L, Yang H, Wang J, Sun J, Wang Z. Integration of multiple genomic and phenotype data to infer novel miRNA-disease associations. PLoS One, 2016, 11(2): e0148521
Carthew R W, Sontheimer E J. Origins and mechanisms of miRNAs and siRNAs. Cell, 2009, 136(4): 642–655
Peng Y, Liu Y, Chen X. Bioinformatics analysis reveals functions of MicroRNAs in rice under the drought stress. Current Bioinformatics, 2020, 15(8): 927–936
Roehle A, Hoefig K P, Repsilber D, Thorns C, Ziepert M, Wesche K O, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, Matolcsy A, Bernd H W, Reiniger L, Merz H, Feller A C. MicroRNA signatures characterize diffuse large B - cell lymphomas and follicular lymphomas. British Journal of Haematology, 2008, 142(5): 732–744
Cogswell J P, Ward J, Taylor I A, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha R K, Richardson J C, Saunders A M, Roses A D, Richards C A. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. Journal of Alzheimer’s Disease, 2008, 14(1): 27–41
Caporali A, Meloni M, Völlenkle C, Bonci D, Sala-Newby G B, Addis R, Spinetti G, Losa S, Masson R, Baker A H, Agami R, Le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation, 2011, 123(3): 282–291
Hu Y, Zhang Y, Zhang H, Gao S, Wang L, Wang T, Han Z, Sun B, Liu G. Cognitive performance protects against Alzheimer’s disease independently of educational attainment and intelligence. Molecular Psychiatry, 2022, 27(10): 4297–4306
Anonymous. 2021 Alzheimer’s disease facts and figures. Alzheimer’s & Dement, 2021, 17(3): 327–406
Hu Y, Sun J, Zhang Y, Zhang H, Gao S, Wang T, Han Z, Wang L, Sun B L, Liu G. rs1990622 variant associates with Alzheimer’s disease and regulates TMEM106B expression in human brain tissues. BMC Medicine, 2021, 19(1): 11
Hu Y, Zhang H, Liu B, Gao S, Wang T, Han Z, International Genomics of Alzheimer’s Project (IGAP), Ji X, Liu G. rs34331204 regulates TSPAN13 expression and contributes to Alzheimer’s disease with sex differences. Brain, 2020, 143(11): e95
Bhaumik D, Scott G K, Schokrpur S, Patil C K, Campisi J, Benz C C. Expression of microRNA-146 suppresses NF-kB activity with reduction of metastatic potential in breast cancer cells. Oncogene, 2008, 27(42): 5643–5647
Wang N, Li Y, Liu S, Gao L, Liu C, Bao X, Xue P. Analysis and validation of differentially expressed MicroRNAs with their target genes involved in GLP-1RA facilitated osteogenesis. Current Bioinformatics, 2021, 16(7): 928–942
Hu Y, Qiu S, Cheng L. Integration of multiple-Omics data to analyze the population-specific differences for coronary artery disease. Computational and Mathematical Methods in Medicine, 2021, 2021: 7036592
Hu Y, Zhang Y, Zhang H, Gao S, Wang L, Wang T, Han Z, International Genomics of Alzheimer’s Project (IGAP), Liu G. Mendelian randomization highlights causal association between genetically increased C-reactive protein levels and reduced Alzheimer’s disease risk. Alzheimer’s & Dement, 2022, 18(10): 2003–2006
Tang W, Wan S, Yang Z, Teschendorff A E, Zou Q. Tumor origin detection with tissue-specific miRNA and DNA methylation markers. Bioinformatics, 2018, 34(3): 398–406
Sarkar J P, Saha I, Sarkar A, Maulik U. Machine learning integrated ensemble of feature selection methods followed by survival analysis for predicting breast cancer subtype specific miRNA biomarkers. Computers in Biology and Medicine, 2021, 131: 104244
Zhu Q, Fan Y, Pan X. Fusing multiple biological networks to effectively predict miRNA-disease associations. Current Bioinformatics, 2021, 16(3): 371–384
Zhang Y, Duan G, Yan C, Yi H, Wu F X, Wang J. MDAPlatform: a component-based platform for constructing and assessing miRNA-disease association prediction methods. Current Bioinformatics, 2021, 16(5): 710–721
Chen X, Zhu C C, Yin J. Ensemble of decision tree reveals potential miRNA-disease associations. PLoS Computational Biology, 2019, 15(7): e1007209
Fu H, Huang F, Liu X, Qiu Y, Zhang W. MVGCN: data integration through multi-view graph convolutional network for predicting links in biomedical bipartite networks. Bioinformatics, 2022, 38(2): 426–434
Zhang G, Li M, Deng H, Xu X, Liu X, Zhang W. SGNNMD: signed graph neural network for predicting deregulation types of miRNA-disease associations. Briefings in Bioinformatics, 2022, 23(1): bbab464
Huang F, Yue X, Xiong Z, Yu Z, Liu S, Zhang W. Tensor decomposition with relational constraints for predicting multiple types of microRNA-disease associations. Briefings in Bioinformatics, 2021, 22(3): bbaa140
Lu X, Gao Y, Zhu Z, Ding L, Wang X, Liu F, Li J. A constrained probabilistic matrix decomposition method for predicting miRNA-disease associations. Current Bioinformatics, 2021, 16(4): 524–533
Lan W, Dong Y, Chen Q, Liu J, Wang J, Chen Y P P, Pan S. IGNSCDA: predicting CircRNA-disease associations based on improved graph convolutional network and negative sampling. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, 19(6): 3530–3538
Peng W, Che Z, Dai W, Wei S, Lan W. Predicting miRNA-disease associations from miRNA-gene-disease heterogeneous network with multi-relational graph convolutional network model. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2022, doi: https://doi.org/10.1109/TCBB.2022.3187739
Chen X, Yan C, Zhang X, You Z H, Deng L, Liu Y, Zhang Y, Dai Q. WBSMDA: within and between score for MiRNA-disease association prediction. Scientific Reports, 2016, 6(1): 21106
Chen X, Yin J, Qu J, Huang L. MDHGI: matrix decomposition and heterogeneous graph inference for miRNA-disease association prediction. PLoS Computational Biology, 2018, 14(8): e1006418
Ding Y, Jiang L, Tang J, Guo F. Identification of human microRNA-disease association via hypergraph embedded bipartite local model. Computational Biology and Chemistry, 2020, 89: 107369
Chen X, Wang L, Qu J, Guan N N, Li J Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics, 2018, 34(24): 4256–4265
Chen X, Sun L G, Zhao Y. NCMCMDA: miRNA-disease association prediction through neighborhood constraint matrix completion. Briefings in Bioinformatics, 2021, 22(1): 485–496
Fu L, Peng Q. A deep ensemble model to predict miRNA-disease association. Scientific Reports, 2017, 7(1): 14482
Zeng X, Ding N, Rodríguez-Patón A, Zou Q. Probability-based collaborative filtering model for predicting gene–disease associations. BMC Medical Genomics, 2017, 10(S5): 76
Zeng X, Liu L, Lü L Y, Zou Q. Prediction of potential disease-associated microRNAs using structural perturbation method. Bioinformatics, 2018, 34(14): 2425–2432
Zeng X, Wang W, Deng G, Bing J, Zou Q. Prediction of potential disease-associated MicroRNAs by using neural networks. Molecular Therapy Nucleic Acids, 2019, 16: 566–575
Chen X, Liu M X, Yan G Y. RWRMDA: predicting novel human microRNA-disease associations. Molecular BioSystems, 2012, 8(10): 2792–2798
Van Laarhoven T, Nabuurs S B, Marchiori E. Gaussian interaction profile kernels for predicting drug-target interaction. Bioinformatics, 2011, 27(21): 3036–3043
Gu C, Liao B, Li X, Li K. Network consistency projection for human miRNA-disease associations inference. Scientific Reports, 2016, 6: 36054
Tiwari P, Dehdashti S, Obeid A K, Marttinen P, Bruza P. Kernel method based on non-linear coherent states in quantum feature space. Journal of Physics A: Mathematical and Theoretical, 2022, 55(35): 355301
Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. HMDD v2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Research, 2014, 42(D1): D1070–D1074
Chen X, Li T H, Zhao Y, Wang C C, Zhu C C. Deep-belief network for predicting potential miRNA-disease associations. Briefings in Bioinformatics, 2021, 22(3): bbaa186
Wang C C, Li T H, Huang L, Chen X. Prediction of potential miRNA-disease associations based on stacked autoencoder. Briefings in Bioinformatics, 2022, 23(2): bbac021
Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 2014, 42(D1): D68–D73
Wang D, Wang J, Lu M, Song F, Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics, 2010, 26(13): 1644–1650
Zhu C C, Wang C C, Zhao Y, Zuo M, Chen X. Identification of miRNA-disease associations via multiple information integration with Bayesian ranking. Briefings in Bioinformatics, 2021, 22(6): bbab302
Zhao Y, Chen X, Yin J. Adaptive boosting-based computational model for predicting potential miRNA-disease associations. Bioinformatics, 2019, 35(22): 4730–4738
Lowe H J, Barnett G O. Understanding and using the medical subject headings (MeSH) vocabulary to perform literature searches. JAMA, 1994, 271(14): 1103–1108
Luo J, Xiao Q, Liang C, Ding P. Predicting MicroRNA-disease associations using Kronecker regularized least squares based on heterogeneous omics data. IEEE Access, 2017, 5: 2503–2513
Lan W, Wang J, Li M, Liu J, Wu F X, Pan Y. Predicting microRNA-disease associations based on improved microRNA and disease similarities. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2018, 15(6): 1774–1782
Lee I, Blom U M, Wang P I, Shim J E, Marcotte E M. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Research, 2011, 21(7): 1109–1121
Cheng L, Wang G, Li J, Zhang T, Xu P, Wang Y. SIDD: a semantically integrated database towards a global view of human disease. PLoS One, 2013, 8(10): e75504
Gretton A, Bousquet O, Smola A, Schölkopf B. Measuring statistical dependence with Hilbert-Schmidt norms. In: Proceedings of the 16th International Conference on Algorithmic Learning Theory. 2005: 63–77
Wang T, Li W. Kernel learning and optimization with Hilbert-Schmidt independence criterion. International Journal of Machine Learning and Cybernetics, 2018, 9(10): 1707–1717
Xuan J, Lu J, Yan Z, Zhang G. Bayesian deep reinforcement learning via deep kernel learning. International Journal of Computational Intelligence Systems, 2018, 12(1): 164–171
Wang T, Lu J, Zhang G. Two-stage fuzzy multiple kernel learning based on Hilbert-Schmidt independence criterion. IEEE Transactions on Fuzzy Systems, 2018, 26(6): 3703–3714
Gönen M, Alpaydin E. Multiple kernel learning algorithms. The Journal of Machine Learning Research, 2011, 12: 2211–2268
Jiang L, Ding Y, Tang J, Guo F. MDA-SKF: similarity kernel fusion for accurately discovering miRNA-disease association. Frontiers in Genetics, 2018, 9: 618
Zhou D, Bousquet O, Lal T N, Weston J, Schölkopf B. Learning with local and global consistency. In: Proceedings of the 16th International Conference on Neural Information Processing Systems. 2003: 321–328
Zhu X, Ghahramani Z, Lafferty J. Semi-supervised learning using Gaussian fields and harmonic functions. In: Proceedings of the 20th International Conference on Machine Learning (ICML-03). 2003: 912–919
Raymond R, Kashima H. Fast and scalable algorithms for semi-supervised link prediction on static and dynamic graphs. In: Proceedings of 2010 European Conference on Machine Learning and Knowledge Discovery in Databases. 2010: 131–147
Laub A J. Matrix Analysis for Scientists and Engineers. Philadelphia: SIAM, 2005
Kashima H, Kato T, Yamanishi Y, Sugiyama M, Tsuda K. Link propagation: a fast semi-supervised learning algorithm for link prediction. In: Proceedings of the 9th SIAM International Conference on Data Mining. 2009: 1093–1104
Golub G H, Hoffman A, Stewart G W. A generalization of the Eckart-Young-Mirsky matrix approximation theorem. Linear Algebra and its Applications, 1987, 88–89: 317–327
Bishop C M, Nasrabadi N M. Pattern Recognition and Machine Learning. New York: Springer, 2006
Vishwanathan S V N, Borgwardt K M, Schraudolph N N. Fast computation of graph kernels. In: Proceedings of the 19th International Conference on Neural Information Processing Systems. 2006
Jiang L, Xiao Y, Ding Y, Tang J, Guo F. FKL-Spa-LapRLS: an accurate method for identifying human microRNA-disease association. BMC Genomics, 2018, 19(S10): 911
Ding Y, Tiwari P, Zou Q, Guo F, Pandey H M. C-loss based higher order fuzzy inference systems for identifying DNA N4-methylcytosine sites. IEEE Transactions on Fuzzy Systems, 2022, 30(11): 4754–4765
Chen X, Xie D, Wang L, Zhao Q, You Z H, Liu H. BNPMDA: bipartite network projection for MiRNA-disease association prediction. Bioinformatics, 2018, 34(18): 3178–3186
Cristianini N, Shawe-Taylor J, Elisseeff A, Kandola J. On kernel-target alignment. In: Proceedings of the 14th International Conference on Neural Information Processing Systems. 2001: 367–373
Cortes C, Mohri M, Rostamizadeh A. Algorithms for learning kernels based on centered alignment. The Journal of Machine Learning Research, 2012, 13(1): 795–828
Lu Y, Wang L, Lu J, Yang J, Shen C. Multiple kernel clustering based on centered kernel alignment. Pattern Recognition, 2014, 47(11): 3656–3664
Hu J, Li Y, Zhang M, Yang X, Shen H B, Yu D J. Predicting protein-DNA binding residues by weightedly combining sequence-based features and boosting multiple SVMs. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2017, 14(6): 1389–1398
Wang H, Tang J, Ding Y, Guo F. Exploring associations of non-coding RNAs in human diseases via three-matrix factorization with hypergraph-regular terms on center kernel alignment. Briefings in Bioinformatics, 2021, 22(5): bbaa409
Chen X, Huang L. LRSSLMDA: laplacian regularized sparse subspace learning for MiRNA-disease association prediction. PLoS Computational Biology, 2017, 13(12): e1005912
You Z H, Huang Z A, Zhu Z, Yan G Y, Li Z W, Wen Z, Chen X. PBMDA: a novel and effective path-based computational model for miRNA-disease association prediction. PLoS Computational Biology, 2017, 13(3): e1005455
Li J Q, Rong Z H, Chen X, Yan G Y, You Z H. MCMDA: matrix completion for MiRNA-disease association prediction. Oncotarget, 2017, 8(13): 21187–21199
Chen X, Yan G Y. Semi-supervised learning for potential human microRNA-disease associations inference. Scientific Reports, 2014, 4: 5501
Xuan P, Han K, Guo M, Guo Y, Li J, Ding J, Liu Y, Dai Q, Li J, Teng Z, Huang Y. Prediction of microRNAs associated with human diseases based on weighted k most similar neighbors. PLoS One, 2013, 8(8): e70204
Chen X, Xie D, Zhao Q, You Z H. MicroRNAs and complex diseases: from experimental results to computational models. Briefings in Bioinformatics, 2019, 20(2): 515–539
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 62072385, 62172076, and U22A2038), the Municipal Government of Quzhou (2022D040), and the Zhejiang Provincial Natural Science Foundation of China (No. LY23F020003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yizheng Wang is a postgraduate in the Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China, China. He received the Bachelor of Engineering degree in computer science and technology from Yanshan University, China in 2022. His research interests include bioinformatics and machine learning.
Xin Zhang is a deputy chief physician of Beidahuang Industry Group General Hospital, China. He graduated from Harbin Medical University, China in 2006, and his research direction is basic medicine and lung cancer.
Ying Ju received her PhD degree in Biomedical Engineering from Xi’an Jiaotong University, China. She is an associate professor with the Department of Computer Science, Xiamen University, China. She has published more than 15 papers in journal and conference. Her main research interest is biomedical engineering.
Qing Liu is a chief physician of Department of Anesthesiology, Hospital (T.C.M) Affiliated to Southwest Medical University, China. He received his master’s degree in Medicine in 2004, and his research interest is mechanism of neuropathic pain.
Quan Zou received the BSc, MSc, and the PhD degrees in computer science from Harbin Institute of Technology, China in 2004, 2007 and 2009, respectively. He is currently a professor in the Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China. His research is in the areas of bioinformatics, machine learning and parallel computing. Several related works have been published by Science, Briefings in Bioinformatics, Bioinformatics, etc. Google scholar showed that his more than 100 papers have been cited more than 16000 times. He is the editor-in-chief of Current Bioinformatics and Computers in Biology and Medicine. He was selected as one of the Clarivate Analytics Highly Cited Researchers in 2018–2022.
Yazhou Zhang received PhD degree in computer applications technology from Tianjin University, China in 2020. He has published more than 35 papers, including CCF ranking A/B conference papers (e.g., IJCAI, EMNLP, CIKM, NAACL) and top journal papers (e.g., IEEE Trans. on Fuzzy System, Information Fusion, ACM Trans. on Internet Technology, Theoretical Computer Science, Neural Networks).
Yijie Ding received the PhD degree in computer science from the School of Computer Science and Technology, Tianjin University, China in 2018. He is currently an Associate Professor with the Yangtze Delta Region Institute, University of Electronic Science and Technology of China, China. His research interests include bioinformatics and machine learning. Several related works have been published by Briefings in Bioinformatics, IEEE TFS, IEEE TAI, IEEE/ACM TCBB, IEEE JBHI, Information Sciences, Knowledge-Based Systems, Applied Soft Computing, and Neurocomputing.
Ying Zhang is a chief physician of Department of Anesthesiology, Hospital (T.C.M) Affiliated to Southwest Medical University, China. She is studying for her PhD at Macau University of Science and Technology, China. She received her master’s degree in Medicine in 2011, and her research interest is mechanism of neuropathic pain and protective mechanism of postoperative cognitive function.
Electronic supplementary material
11704_2023_2490_MOESM1_ESM.pdf
Identification of human microRNA-disease association via low-rank approximation-based link propagation and multiple kernel learning
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, X., Ju, Y. et al. Identification of human microRNA-disease association via low-rank approximation-based link propagation and multiple kernel learning. Front. Comput. Sci. 18, 182903 (2024). https://doi.org/10.1007/s11704-023-2490-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11704-023-2490-5