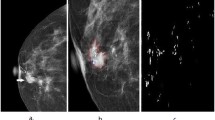
Abstract
Though rare in male, breast cancer is very frequent in women and is of fatal nature. Microcalcification (MC), an early indication of breast cancer, can reduce the mortality rate by many folds if detected and diagnosed at the earlier stage. But, due to its existence in discrete as well as in clustered form, its identification becomes a challenging task. Considering the recent advances and impact of deep learning techniques in biomedical imaging, in this paper, deep learning architectures, U-Net and its modified version: SqueezeU-Net, are proposed for the detection of MC followed by its characterization as benign or malignant. Being fully convolutional network, U-Net and SqueezeU-Net both are independent of dimensions of the input. SqueezeU-Net has less computational complexity while preserving the accuracy of the system due to delay downsampling. The assessment of the proposed model is performed on 1000 mammograms of DDSM dataset and the analysis is extended with data augmentation. A true positive rate (TPR) of 89.83% with 0.42 false positive per image (FPs/I) is obtained for SqueezeU-Net as compared to 84.67% at 0.5 FPs/I by U-Net architecture. The detected MC were classified and achieved an accuracy of 97.30% and area under the ROC curve (AUC) is 0.97 for SqueezeU-Net while the same for U-Net are 93.64% and 0.93 respectively using five-fold cross validation. The results are further compared with other competing schemes of the state-of-the-art where the proposed architectures take an edge over others.






Similar content being viewed by others
References
Akselrod-Ballin, A., Karlinsky, L., Hazan, A., et al.: Deep Learning for Automatic Detection of Abnormal Findings in Breast Mammography, pp. 321–329. Springer International Publishing, Cham (2017)
Astley, S., Gilbert, F.: Computer-aided detection in mammography. Clin. Radiol. 59(5), 390–399 (2004)
Bakkouri, I., Afdel, K.: Multi-scale cnn based on region proposals for efficient breast abnormality recognition. Multimedia Tools and Applications 78(10), 12,939-12,960 (2019)
Basile, T.M.A., Fanizzi, A., Losurdo, L., et al.: Microcalcification detection in full-field digital mammograms: A fully automated computer-aided system. Physica Med. 64, 1–9 (2019)
Bekker, A.J., Greenspan, H., Goldberger, J.: A multi-view deep learning architecture for classification of breast microcalcifications. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 726–730 (2016)
Cai, H., Huang, Q., Rong, W., et al.: Breast microcalcification diagnosis using deep convolutional neural network from digital mammograms. Comput. Math. Methods Med. 2717, 454 (2019)
Carneiro, G., Nascimento, J., Bradley, A.P.: Automated analysis of unregistered multi-view mammograms with deep learning. IEEE Trans. Med. Imaging 36(11), 2355–2365 (2017)
Ferlay, J., Soerjomataram, I., Dikshit, R., et al.: Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 5, E359–E386 (2015)
Greenspan, H., van Ginneken, B., Summers, R.M.: Guest editorial deep learning in medical imaging: Overview and future promise of an exciting new technique. IEEE Trans. Med. Imaging 35(5), 1153–1159 (2016)
Hamidinekoo, A., Denton, E., Rampun, A., et al.: Deep learning in mammography and breast histology, an overview and future trends. Med. Image Anal. 47, 45–67 (2018)
He, K., Sun, J.: Convolutional neural networks at constrained time cost (2014)
Heath, M., Bowyer, K., Kopans, D., et al.: The digital database for screening mammography. In: Proceedings of the Fourth International Workshop on Digital Mammography (2000)
Iandola, F.N., Han, S., Moskewicz, M.W., et al.: Squeezenet: Alexnet-level accuracy with 50x fewer parameters and \(<\)0.5mb model size (2016)
Lu, Z., Carneiro, G., Dhungel, N., et al.: Automated detection of individual micro-calcifications from mammograms using a multi-stage cascade approach (2016)
Ma, Y., Tay, P.C., Adams, R.D., et al.: A novel shape feature to classify microcalcifications. In: 2010 IEEE International Conference on Image Processing, 2010 IEEE International Conference on Image Processing, pp. 2265–2268 (2010)
Mordang, J.J., Janssen, T., Bria, A., et al.: Automatic microcalcification detection in multi-vendor mammography using convolutional neural networks. pp. 35–42 (2016)
O’Shea, K., Nash, R.: An introduction to convolutional neural networks (2015)
Pisano, E.D., Gatsonis, C., Hendrick, E., et al.: Diagnostic performance of digital versus film mammography for breast-cancer screening. N. Engl. J. Med. 353(17), 1773–1783 (2005). (PMID: 16169887)
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional networks for biomedical image segmentation (2015)
Scherer, D., Müller, A., Behnke, S.: Evaluation of pooling operations in convolutional architectures for object recognition. In: Diamantaras, K., Duch, W., Iliadis, L.S. (eds.) Artifi Neural Netw. ICANN 2010, pp. 92–101. Springer, Berlin (2010)
Sickles, E.A.: Breast calcifications: mammographic evaluation. Radiology 160(2), 289–293 (1986). (PMID: 3726103)
Sung, H., Ferlay, J., Siegel, R.L., et al.: Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021). https://doi.org/10.3322/caac.21660
Taylor, L., Nitschke, G.: Improving deep learning using generic data augmentation (2017)
Wang, J., Yang, Y.: A context-sensitive deep learning approach for microcalcification detection in mammograms. Pattern Recogn. 78, 12–22 (2018)
Wang, J., Nishikawa, R.M.: Global detection approach for clustered microcalcifications in mammograms using a deep learning network. J. Med. Imaging 4(2), 297–303 (2017)
Xi, P., Shu, C., Goubran, R.: Abnormality detection in mammography using deep convolutional neural networks. In: 2018 IEEE International Symposium on Medical Measurements and Applications (MeMeA), pp. 1–6 (2018)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kulkarni, S., Rabidas, R. SqueezeU-Net-based detection and diagnosis of microcalcification in mammograms. SIViP 17, 435–443 (2023). https://doi.org/10.1007/s11760-022-02240-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02240-0