Abstract
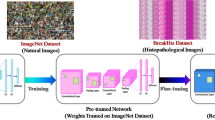
Breast cancer (BC) is a massive health problem and a deadly disease, killing millions of people every year. Computerized approaches for automated malignant BC detection can efficiently help in reducing the manual workload of pathologists and making diagnosis more scalable and less prone to errors. In this paper, we present two systems to diagnose breast cancer from single and multi-magnification histopathological images. The first proposed system utilizes a pre-trained DenseNet201 CNN architecture and fine-tuned over the publicly available BreakHis dataset and classifies histopathological images of specific magnification factors into one of the benign or malignant classes. The second system consists of four subsystems, each corresponding to one of the magnifications, and is trained only by related magnification images. Afterwards, the results obtained from these four subsystems are fused together to make the final decision. Several experiments on BreakHis dataset demonstrate that the proposed systems outperform the state-of-the-art approaches, in all cases.



Similar content being viewed by others
References
Boyle, P., Levin, B. (Eds).: World Cancer Report. Lyon: IARC, 2008. [Online]. Available: https://publications.iarc.fr/_publications/media/download/4083/0d98fe6c30034ada27810fc3049ece916392c163.pdf (2008)
Joy, J.E., Penhoet, E.E., Petitti, D.B., Institute of Medicine (US) and National Research Council (US) Committee on New Approaches to Early Detection and Diagnosis of Breast Cancer (Eds.).: Saving Women's Lives: Strategies for Improving Breast Cancer Detection and Diagnosis. National Academies Press (US). (2005)
Gurcan, M.N., Boucheron, L.E., Can, A., Madabhushi, A., Rajpoot, N.M., Yener, B.: Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2, 147–171 (2009)
Désir, C., Petitjean, C., Heutte, L., Salaun, M., Thiberville, L.: Classification of endomicroscopic images of the lung based on random subwindows and extra-trees. IEEE Trans. Biomed. Eng. 59(9), 2677–2683 (2012)
Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., Sánchez, C.I.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.:. Breast cancer histopatho- logical image classification using convolutional neural networks. In: international joint conference on neural networks (IJCNN), pp. 2560–2567. IEEE. (2016)
LeCun, Y., Boser, B.E., Denker, J.S., Henderson, D., Howard, R.E., Hubbard, W.E., Jackel, L.D.: Handwritten digit recognition with a back-propagation network. Adv. Neural Inf. Process. Syst. 2, 396–404 (1990)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: 2012 AlexNet. Adv. Neural Inf. Process. Syst., 1–9. (2012)
Samah, A.A., Fauzi, M.F.A., Mansor, S.: Classification of benign and malignant tumors in histopathology images. In: IEEE international conference on signal and image processing applications (ICSIPA), pp. 102- 106. IEEE. (2017)
Kahya, M.A., Al-Hayani, W., Algamal, Z.Y.: Classification of breast cancer histopathology images based on adaptive sparse support vector machine. J. Appl. Math. Bioinf. 7(1), 49 (2017)
Liao, C., Li, S., Luo, Z.: Gene selection using wilcoxon rank sum test and support vector machine for cancer classification. In: international conference on computational and information science, pp. 57–66. Springer, Berlin, Heidelberg. (2006)
Benhammou, Y., Achchab, B., Herrera, F., Tabik, S.: BreakHis based breast cancer automatic diagnosis using deep learning: taxonomy, survey and insights. Neurocomputing 375, 9–24 (2020)
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., Rabinovich, A.: Going deeper with convolutions. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1–9. (2015)
Benhammou, Y., Tabik, S., Achchab, B., Herrera, F.: A first study exploring the performance of the state-of-the art CNN model in the problem of breast cancer. In: proceedings of the international conference on learning and optimization algorithms: theory and applications, pp. 1–6. Morocco (2018)
Donahue, J., Jia, Y., Vinyals, O., Hoffman, J., Zhang, N., Tzeng, E., Darrell, T.: Decaf: A deep convolutional activation feature for generic visual recognition. In: international conference on machine learning, pp. 647–655. (2014)
Deniz, E., Şengür, A., Kadiroğlu, Z., Guo, Y., Bajaj, V., Budak, Ü.: Transfer learning based histopathologic image classification for breast cancer detection. Health Inf. Sci. Syst. 6(1), 1–7 (2018)
Badejo, J.A., Adetiba, E., Akinrinmade, A., Akanle, M.B.: Medical image classification with hand-designed or machine-designed texture descriptors: a performance evaluation. In: international conference on bioinformatics and biomedical engineering, pp. 266–275. Springer, Cham. (2018)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2015)
Taheri, S., Toygar, Ö.: On the use of DAG-CNN architecture for age estimation with multi-stage features fusion. Neurocomputing 329, 300–310 (2019)
Golrizkhatami, Z., Acan, A.: ECG classification using three-level fusion of different feature descriptors. Expert Syst. Appl. 114, 54–64 (2018)
Deng, J., Dong, W., Socher, R., Li, L. J., Li, K., Fei-Fei, L.: Imagenet: A large-scale hierarchical image database. In: 2009 IEEE conference on computer vision and pattern recognition, pp. 248–255. IEEE (2009)
Gupta, V., Bhavsar, A.: An integrated multi-scale model for breast cancer histopathological image classification with joint colour-texture features. In: international conference on computer analysis of images and patterns, pp. 354–366. Springer, Cham. (2017)
Taheri, S., Toygar, Ö.: Multi-stage age estimation using two level fusions of handcrafted and learned features on facial images. IET Biometrics 8(2), 124–133 (2018)
Simonyan, K., Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. (2014)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. Computer Science. (2015)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4700–4708. (2017)
Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1251–1258. IEEE (2017)
Zoph, B., Vasudevan, V., Shlens, J., Le, Q.V.: Learning transferable architectures for scalable image recognition. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp. 8697–8710. (2018)
Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi, A.: Inception-v4, inception-resnet and the impact of residual connections on learning. In: proceedings of the AAAI conference on artificial intelligence, vol 31, No. 1.(2017)
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.C.: Mobilenetv2: Inverted residuals and linear bottlenecks. In: proceedings of the IEEE conference on computer vision and pattern recognition, pp. 4510–4520. (2018)
Spanhol, F.A., Oliveira, L.S., Cavalin, P.R., Petitjean, C., Heutte, L.:. Deep features for breast cancer histopathological image classification. In: IEEE international conference on systems, man, and cybernetics (SMC), pp. 1868–1873. IEEE. (2017)
Song, Y., Zou, J.J., Chang, H., Cai, W.: Adapting fisher vectors for histopathology image classification. In: 2017 IEEE 14th international symposium on biomedical imaging (ISBI 2017), pp. 600–603. IEEE. (2017)
Kumar, K., Rao, A. C. S.: Breast cancer classification of image using convolutional neural network. In: 4th international conference on recent advances in information technology (RAIT), pp. 1–6. IEEE. (2018)
Sanchez-Morillo, D., González, J., García-Rojo, M., Ortega, J.: Classification of breast cancer histopathological images using KAZE features. In: international conference on bioinformatics and biomedical engineering, pp. 276–286. Springer, Cham. (2018)
Sudharshan, P.J., Petitjean, C., Spanhol, F., Oliveira, L.E., Heutte, L., Honeine, P.: Multiple instance learning for histopathological breast cancer image classification. Expert Syst. Appl. 117, 103–111 (2019)
Song, Y., Chang, H., Huang, H., Cai, W.: Supervised intra-embedding of fisher vectors for histopathology image classification. In: international conference on medical image computing and computer-assisted intervention, pp. 99–106. Springer, Cham. (2017)
Bayramoglu, N., Kannala, J., Heikkilä, J.: Deep learning for magnification independent breast cancer histopathology image classification. In: 23rd international conference on pattern recognition (ICPR), pp. 2440–2445. IEEE. (2016)
Gupta, V., Bhavsar, A.: Breast cancer histopathological image classification: is magnification important?. In: proceedings of the IEEE conference on computer vision and pattern recognition workshops, pp. 17–24. (2017)
Zhang, Z., Chen, B., Xu, S., Chen, G., Xie, J.: A novel voting convergent difference neural network for diagnosing breast cancer. Neurocomputing 437, 339–350 (2021)
Boumaraf, S., Liu, X., Wan, Y., Zheng, Z., Ferkous, C., Ma, X., Bardou, D.: Conventional machine learning versus deep learning for magnification dependent histopathological breast cancer image classification: a comparative study with visual explanation. Diagnostics 11(3), 528 (2021)
Hameed, Z., Zahia, S., Garcia-Zapirain, B., Javier Aguirre, J., María Vanegas, A.: Breast cancer histopathology image classification using an ensemble of deep learning models. Sensors 20(16), 4373 (2020)
Sharma, S., Mehra, R.: Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images—a comparative insight. J. Digit. Imaging 33(3), 632–654 (2020)
Zhang, Z., Chen, G., Yang, S.: Ensemble support vector recurrent neural network for brain signal detection. IEEE Trans. Neural Netw. Learn. Syst. (2021). https://doi.org/10.1109/TNNLS.2021.3083710
Zhang, Z., Chen, B., Sun, J., Luo, Y.: A bagging dynamic deep learning network for diagnosing COVID-19. Sci. Rep. 11, 16280 (2021)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Taheri, S., Golrizkhatami, Z. Magnification-specific and magnification-independent classification of breast cancer histopathological image using deep learning approaches. SIViP 17, 583–591 (2023). https://doi.org/10.1007/s11760-022-02263-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02263-7