Abstract
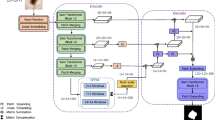
Accurate automatic segmentation of medical images is required in computer-aided diagnosis systems in clinical medicine. Convolutional neural networks (CNNs) based on U-shaped structures are widely used in medical image segmentation tasks. However, due to the intrinsic locality of the convolution operation, it is difficult for CNN-based approaches to learn the global information and long-range semantic information interactions using Swin-Unet. However, we find that UNet and Swin-Unet have the worst segmentation performance on small masses. To remedy this problem, this paper presents an end-to-end depthwise separable U-shaped convolution network with a large convolution kernel (DS-UNeXt) for the medical image segmentation of computed tomography (CT) images and magnetic resonance images (MRIs). Our network has a larger receptive field to extract features, which is useful for boosting the performance of multiscale medical segmentations. In DS-UNeXt, parallel depthwise separable spatial pooling (PDSP) is proposed to aggregate the global information. PDSP consists of multiple parallel depthwise separable convolutions to enhance the high-level semantic features. The proposed DS-UNeXt achieves Dice indices of 80.65% and 90.88% on the synapse for the multiorgan segmentation dataset and the automatic cardiac diagnosis challenge (ACDC) dataset, respectively. Moreover, extensive experiments show that DS-UNeXt transcends several state-of-the-art segmentation networks.




Similar content being viewed by others
Availability of data and materials
We conducted experiments on three datasets, including the Synapse for multiorgan CT segmentation dataset and ACDC dataset. The Synapse for multiorgan CT segmentation dataset can be found in https://www.synapse.org/#!Synapse:syn3193805/wiki/217789. The ACDC dataset can be found in https://acdc.creatis.insa-lyon.fr/description/databases.html.
References
Sun, S., Liu, Y., Bai, N., et al.: Attentionanatomy: A unified framework for whole-body organs at risk segmentation using multiple partially annotated datasets. In: Proceedings of the IEEE International Symposium on Biomedical Imaging, pp. 1–5 (2020)
Tang, H., Zhang, C., Xie, X.: Automatic pulmonary lobe segmentation using deep learning. In: Proceedings of the IEEE International Symposium on Biomedical Imaging, pp. 1225–1228 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241 (2015)
Isensee, F., Jaeger, P.F., Kohl, S.A., et al.: nnUNet: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021)
Asgari Taghanaki, S., Abhishek, K., Cohen, J.P., et al.: Deep semantic segmentation of natural and medical images: a review. Artif. Intell. Rev. 54, 137–178 (2021)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., et al.: 3D UNet: learning dense volumetric segmentation from sparse annotation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 424–432 (2016)
Xiao, X., Lian, S., Luo, Z., et al.: Weighted res-unet for high-quality retina vessel segmentation. In: Proceedings of the International Conference on Information Technology in Medicine and Education, pp..327–331 (2018)
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., et al.: UNet++: a nested UNet architecture for medical image segmentation. In: Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, pp. 3–11 (2018)
Oktay O., Schlemper, J., Folgoc, L.L., et al.: Attention UNet: learning where to look for the pancreas. arXiv:1804.03999 (2018)
Huang, H., Lin, L., Tong, R., et al.: UNet 3+: a full-scale connected UNet for medical image segmentation. In: Proceedings of the ICASSP 2020–2020 IEEE International Conference on Acoustics, pp. 1055–1059 (2020)
Karimi Jafarbigloo, S., Danyali, H.: Nuclear atypia grading in breast cancer histopathological images based on CNN feature extraction and LSTM classification. CAAI Trans. Intell. Technol. 6, 426–439 (2021)
Jia, Y., Wang, H., Chen, W., et al.: An attention-based cascade R-CNN model for sternum fracture detection in X-ray images. CAAI Trans. Intell. Technol. (2022). https://doi.org/10.1049/cit2.12072
Vaswani, A., Shazeer, N., Parmar, N., et al.: Attention is all you need. In: Proceedings of the Advances in Neural Information Processing Systems, pp. 4–9 (2017)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., et al.: An image is worth 16 × 16 words: transformers for image recognition at scale. arXiv:2010.11929 (2020)
Chen, J., Lu, Y., Yu, Q., et al.: TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306 (2021)
Zhou, H. Y., Guo, J., Zhang, Y., et al.: nnformer: interleaved transformer for volumetric segmentation. arXiv:2109.03201 (2021)
Hatamizadeh, A., Tang, Y., Nath, V., et al.: Unetr: transformers for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584 (2022)
Jun, E., Jeong, S, Heo, D.W., et al.: Medical transformer: universal brain encoder for 3D MRI analysis. arXiv:2104.13633 (2021)
He, S., Grant, P.E., Ou, Y.: Global-local transformer for brain age estimation. IEEE Trans. Med. Imaging 41, 213–224 (2021)
Costa, G.S.S., Paiva, A.C., Junior, G.B., et al.: COVID-19 automatic diagnosis with CT images using the novel transformer architecture. In: Anais do XXI simpósio brasileiro de computação aplicada à saúde, pp. 293–301 (2021)
Liu, Z., Lin, Y., Cao, Y., et al.: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021)
Cao, H., Wang, Y., Chen, J., et al.: Swin-Unet: Unet-like pure transformer for medical image segmentation. arXiv:2105.05537 (2021)
Lin, A., Chen, B., Xu, J., et al.: Ds-transunet: dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 71, 1–15 (2022)
Liu, Z., Mao, H., Wu, C.Y., et al.: A convnet for the 2020s. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 11976–11986 (2022)
Howard, A.G., Zhu, M., Chen, B., et al.: MobileNets: efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv:1704.04861 (2017)
Tsai, A., Yezzi, A., Wells, W., et al.: A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med. Imaging 22, 137–154 (2003)
Held, K., Kops, E.R., Krause, B.J., et al.: Markov random field segmentation of brain MR images. IEEE Trans. Med. Imaging 16, 878–886 (1997)
Patil, D.D., Deore, S.G.: Medical image segmentation: a review. Int. J. Comput. Sci. Mobile Comput. 2(1), 22–27 (2013)
Cao, L., Liang Y., Lv, W., et al.: Relating brain structure images to personality characteristics using 3D convolution neural network. In: Proceedings of the CAAI Transactions on Intelligence Technology, vol. 6(3), pp. 338–346 (2021)
Cao, Y., Liu, S., Peng, Y., et al.: DenseUNet: densely connected UNet for electron microscopy image segmentation. IET Image Proc. 14, 2682–2689 (2020)
Zhao, H., Qiu, X., Lu, W., Huang, H., et al.: High-quality retinal vessel segmentation using generative adversarial network with a large receptive field. Int. J. Imaging Syst. Technol. 30(3), 828–842 (2020)
Chen, L., Bentley, P., Mori, K., et al.: DRINet for medical image segmentation. IEEE Trans. Med. Imaging 37(11), 2453–2462 (2018)
Milletari, F., Nassir N., Ahmadi, S.A.: V-net: fully convolutional neural networks for volumetric medical image segmentation. In: Proceedings of the 2016 Fourth International Conference on 3D Vision, pp. 565–571 (2016)
Devlin, J., Chang, M.W., Lee, K., et al.: Bert: pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805 (2018)
Zhang, Y., Du, T., Sun, Y., et al.: Form 10-q itemization. In: Proceedings of the 30th ACM International Conference on Information Knowledge Management, pp. 4817–4822 (2021)
Chang, Y., Menghan, H., Guangtao, Z., et al.: Transclaw UNet: claw UNet with transformers for medical image segmentation. arXiv:2107.05188 (2021)
Sha, Y., Zhang, Y., Ji, X., et al.: Transformer-UNet: raw image processing with UNet. arXiv:2109.08417 (2021)
Gao, Y., Zhou, M., Metaxas, D.N.: UTNet: a hybrid transformer architecture for medical image segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 61–71 (2021)
Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., et al.: Medical transformer: gated axial-attention for medical image segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 36–46 (2021)
Xie, Y., Zhang, J., Shen, C., et al.: Cotr: efficiently bridging cnn and transformer for 3d medical image segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, pp. 171–180 (2021)
Tang, Y., Yang, D., Li, W., et al.: A. Self-supervised pre-training of swin transformers for 3d medical image analysis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 20730–20740 (2022)
Ba, J.L., Kiros, J.R., Hinton, G.E. Layer normalization. arXiv:1607.06450 (2016)
Ioffe, S.: Batch renormalization: towards reducing minibatch dependence in batch-normalized models. In: Proceedings of the Advances in Neural Information Processing Systems, p. 30 (2017)
Nair, V., Hinton, G.E.: Rectified linear units improve restricted Boltzmann machines. In: Proceedings of the 27th International Conference on Machine Learning, pp. 21–24 (2010)
Chen, L.C., Papandreou, G., Kokkinos, I., et al.: Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 40, 834–848 (2017)
Xie, S., Girshick, R., Dollár, P., et al.: Aggregated residual transformations for deep neural networks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 1492–1500 (2017)
Sandler, M., Howard, A., Zhu, M., et al.: Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
Hendrycks, D., Kevin, G.: Gaussian error linear units (gelus). arXiv:1606.08415 (2016)
Fu, S., Lu, Y., Wang, Y., et al.: Domain adaptive relational reasoning for 3d multi-organ segmentation. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 656–666 (2020)
Bernard, O., Lalande, A., Zotti, C., et al.: Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans. Med. Imaging 37, 2514–2525 (2018)
Loshchilov, I., Frank, H.: Decoupled weight decay regularization. arXiv:1711.05101 (2017)
Schlemper, J., Oktay, O., Schaap, M., et al.: Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019)
Zhao, H., Shi, J., Qi, X., et al.: Pyramid scene parsing network. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2881–2890 (2017)
Funding
This work was supported by the Natural Science Foundation of Chongqing, China (Grant No. cstc2021jcyj-msxmX0605), and Science and Technology Foundation of Chongqing Education Commission (Grant No. KJQN202001137).
Author information
Authors and Affiliations
Contributions
T.H. and J.C. contributed to conceptualization, methodology, software, 402, validation, formal analysis, investigation, 403 resources, data curation, writing—original draft preparation, writing—review and editing, and visualization. L.J. contributed to supervision, project 405 administration, and funding acquisition. All authors have read and agreed to the published 406 version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Synapse for multiorgan CT segmentation dataset and ACDC dataset belongs to public datasets. The patients involved in the dataset have obtained ethical approval. User can download relevant data for free for research and publish relevant articles. Our study is based on open-source data, so there are no ethical issues.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Huang, T., Chen, J. & Jiang, L. DS-UNeXt: depthwise separable convolution network with large convolutional kernel for medical image segmentation. SIViP 17, 1775–1783 (2023). https://doi.org/10.1007/s11760-022-02388-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02388-9