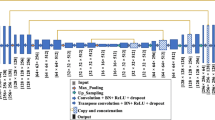
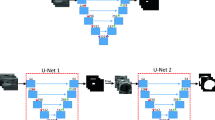
Abstract
Manual segmentation of breast lesions is a tedious and time-consuming task. The existing state-of-the-art studies are evaluated on single modalities, i.e., either on mammogram or ultrasound thereby having limited clinical application. The limitations stated above motivated us to develop a Computer-Aided Mammogram Segmentation (CAMS) system and Computer-Aided Ultrasound Segmentation (CAUS) by using a customized pre-trained AlexNet network to perform semi-automated segmentation of breast lesions in dual-modality. A new real-time dual-modality data of mammogram and ultrasound are developed for conducting this study. Data augmentation is applied to improve diversity of the data. The last layer of the pre-trained network is modified to perform pixel-wise classification of the input test image. The output binary image is obtained using the color map of each test image. Various performance measures and statistical correlation values of paired T test are used for evaluation of the proposed model. The proposed model achieves a Jaccard index of 0.53 for mammogram and 0.63 for ultrasound, respectively. Further, a Dice similarity coefficient of 0.66 for mammogram and 0.76 for ultrasound is achieved using the proposed CAMS and CAUS systems, respectively. Area under curve of the proposed CAUS system is found to be 0.81 which is very close to that of the CAMS, i.e., 0.82. It is concluded that the CAUS system can be useful in breast lesion detection along with CAMS in the routine clinical scenario. Ultrasound being a painless, ionizing radiation-free, and low-cost technology can be useful for breast cancer screening over traditional mammograms, particularly in low settings.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.Availability of data and materials
Data are not available with this manuscript but can be made available on reasonable request after permission of IEC.
References
National Cancer Registry Programme. Three-year report of population based cancer registries: 2012–2014. Chapter10_Printed.pdf (https://ncdirindia.org) (2016). Accessed 10 June 2021
Cheng, H.D., Shan, J., Ju, W., Guo, Y., Zhang, L.: Automated breast cancer detection and classification using ultrasound images: a survey. Pattern Recognit. 43, 299–317 (2010). https://doi.org/10.1016/j.patcog.2009.05.012
Yuan, Y., Giger, M.L., Li, H., Bhooshan, N., Sennett, C.A.: Multimodality computer-aided breast cancer diagnosis with FFDM and DCE-MRI. Acad. Radiol. 17, 1158–1167 (2010). https://doi.org/10.1016/j.acra.2010.04.015
El Atlas, N., El Aroussi, M., Wahbi, M.: Computer-aided breast cancer detection using mammograms: a review. In: WCCS 2014: Second World Conference on Complex Systems (WCCS), pp. 626–631 (2014). https://doi.org/10.1109/ICoCS.2014.7060995
Singh, B.K., Jain, P., Banchhor, S.K., Verma, K.: Performance evaluation of breast lesion detection systems with expert delineations: a comparative investigation on mammographic images. Multimed. Tools Appl. (2019). https://doi.org/10.1007/s11042-019-7570-z
Patel, B.C., Sinha, G.R., Soni, D.: Detection of masses in mammographic breast cancer images using modified histogram based adaptive thresholding (MHAT) method. Int. J. Biomed. Eng. Technol. 29, 134–154 (2019). https://doi.org/10.1504/IJBET.2019.097302
Luo, Y., Liu, L., Huang, Q., Li, X.: A novel segmentation approach combining region-and edge-based information for ultrasound images. Biomed. Res. Int. (2017). https://doi.org/10.1155/2017/9157341
Kumar, V., Webb, J.M., Gregory, A., Denis, M., Meixner, D.D., Bayat, M.M., Whaley, D.H., Fatemi, M., Alizad, A.: Automated and real-time segmentation of suspicious breast masses using convolutional neural network. PLoS ONE. 13(5), e0195816 (2018). https://doi.org/10.1371/journal.pone.0195816
Xie, X., Shi, F., Niu, J., Tang, X.: Breast ultrasound image classification and segmentation using convolutional neural networks. In: Pacific Rim Conference on Multimedia, pp. 200–211. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00764-5_19
Liu, L., Li, K., Qin, W., Wen, T., Li, L., Wu, J., Gu, J.: Automated breast tumor detection and segmentation with a novel computational framework of whole ultrasound images. Med. Biol. Eng. Comput. 56, 183–199 (2018). https://doi.org/10.1007/s11517-017-1770-3
Xu, Y., Wang, Y., Yuan, J., Cheng, Q., Wang, X., Carson, P.L.: Medical breast ultrasound image segmentation by machine learning. Ultrasonics 91, 1–9 (2019). https://doi.org/10.1016/j.ultras.2018.07.006
Panigrahi, L., Verma, K., Singh, B.K.: Ultrasound image segmentation using a novel multi-scale Gaussian kernel fuzzy clustering and multi-scale vector field convolution. Expert Syst. Appl. 115, 486–498 (2019). https://doi.org/10.1016/j.eswa.2018.08.013
Huang, Q., Huang, Y., Luo, Y., Yuan, F., Li, X.: Segmentation of breast ultrasound image with semantic classification of superpixels. Med. Image Anal. 61, 101657 (2020). https://doi.org/10.1016/j.media.2020.101657
Xue, C., Zhu, L., Fu, H., Hu, X., Li, X., Zhang, H., Heng, P.A.: Global guidance network for breast lesion segmentation in ultrasound images. Med. Image Anal. 70, 101989 (2021). https://doi.org/10.1016/j.media.2021.101989
Huang, Q., Miao, Z., Zhou, S., Chang, C., Li, X.: Dense prediction and local fusion of superpixels: a framework for breast anatomy segmentation in ultrasound image with scarce data. IEEE Trans. Instrum. Meas. 70, 1–8 (2021). https://doi.org/10.1109/TIM.2021.3088421
Qiao, M., Liu, C., Li, Z., Zhou, J., Xiao, Q., Zhou, S., Chang, C., Gu, Y., Guo, Y., Wang, Y.: Breast tumor classification based on MRI-US images by disentangling modality features. IEEE J. Biomed. Health Inform. 26(7), 3059–3067 (2022). https://doi.org/10.1109/JBHI.2022.3140236
Pan, S.J., Yang, Q.: A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2009). https://doi.org/10.1109/TKDE.2009.191
Gong, S., Liu, C., Ji, Y., Zhong, B., Li, Y., Dong, H.: Image and video understanding based on deep learning. In: Advanced Image and Video Processing Using MATLAB, pp. 513–553. Springer, Cham (2019). https://doi.org/10.1007/978-3-319-77223-3_14
Heath, M., Bowyer, K., Kopans, D., Moore, R., Kegelmeyer, W.P.: The digital database for screening mammography. In: Yaffe, M.J. (ed.) Proceedings of the Fifth International Workshop on Digital Mammography, pp. 212–218. Medical Physics Publishing (2001).
Heath, M., Bowyer, K., Kopans, D., Kegelmeyer, W.P., Moore, R., Chang, K., MunishKumaran, S.: Current status of the digital database for screening mammography. In: Digital Mammography, Proceedings of the Fourth International Workshop on Digital Mammography, pp. 457–460. Kluwer Academic Publishers (1998)
Al-Dhabyani, W., Gomaa, M., Khaled, H., Fahmy, A.: Dataset of breast ultrasound images. Data Brief. 28, 104863 (2020). https://doi.org/10.1016/j.dib.2019.104863
Online dataset: http://www.onlinemedicalimages.com/index.php/en/site-map
Rodrigues, P.S.: Breast Ultrasound Image, Mendeley Data, v1 (2017). https://doi.org/10.17632/wmy84gzngw.1
Daoud, M.I., Atallah, A.A., Awwad, F., Al-Najjar, M., Alazrai, R.: Automatic superpixel-based segmentation method for breast ultrasound images. Expert Syst. Appl. 121, 78–96 (2019). https://doi.org/10.1016/j.eswa.2018.11.024
Shorten, C., Khoshgoftaar, T.M.: A survey on image data augmentation for deep learning. J. Big Data. 6, 1–48 (2019). https://doi.org/10.1186/s40537-019-0197-0
Zhu, Y., Fu, Z., Fei, J.: An image augmentation method using convolutional network for thyroid nodule classification by transfer learning. In: 2017 3rd IEEE International Conference on Computer and Communications (ICCC), pp. 1819–1823. IEEE (2017). https://doi.org/10.1109/CompComm.2017.8322853
Bowles, D., Quinton, A.: The use of ultrasound in breast cancer screening of asymptomatic women with dense breast tissue: a narrative review. J. Med. Imaging Radiat. Sci. 47, S21–S28 (2016). https://doi.org/10.1016/j.jmir.2016.06.005
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
KA and AR contributed to methodology, software, and writing—original draft. BKS contributed to conceptualization, methodology, and supervision. NKB contributed to conceptualization and supervision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Institutional ethics committee (IEC), NIT Raipur Letter No.: NITRR/IEC/2019/04.
Informed consent
Informed consent was obtained from all the participants in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Atrey, K., Singh, B.K., Roy, A. et al. A dual-modality evaluation of computer-aided breast lesion segmentation in mammogram and ultrasound using customized transfer learning approach. SIViP 17, 1955–1963 (2023). https://doi.org/10.1007/s11760-022-02408-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-022-02408-8