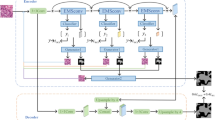
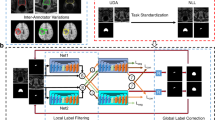
Abstract
Breast cancer, a high-incidence cancer among female, occupies a large incidence of total female patients with cancer. Pathological examination is the gold standard for breast cancer in clinic diagnosis. However, accuracy and efficient diagnosis is challengeable to pathologists for the complex of breast cancer and laborious work. Introducing computer-aid diagnosis (CAD) can relieve laborious work of pathologists and improve diagnosed accuracy for breast cancer. To promote development of CAD methods, we release a large-scale and hematoxylin-eosin (HE) staining dataset of breast cancer for medical image segmentation task, called the breast-cancer image segmentation 5000 (BIS5k). BIS5k contains 5929 images that are divided into training data (5000) and evaluated data (929). All images of BIS5k are collected from clinic cases which include patients with various age and cancer stages. All labels of images are annotated in pixel level for segmentation task and reviewed by pathological professors carefully. Furthermore, we construct a basic instance called breast-cancer segmentation network, BCSNet with a toolkit including comprehensive metrics to demonstrate the usage of BIS5k. Extensive experiments of BCSNet and compared methods provide that developing specific algorithm and constructing dataset are indispensable to promote CAD of pathological diagnosis for breast cancer.




Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Araujo, T., Aresta, G., Castro, E., Rouco, J., Aguiar, P., Eloy, C., Polónia, A., Campilho, A.: Classification of breast cancer histology images using convolutional neural networks. PloS One 12(6), 0177544 (2017)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: Breast cancer histopathological image classification using convolutional neural networks. In: 2016 International Joint Conference on Neural Networks (IJCNN), pp. 2560–2567 (2016)
Spanhol, F.A., Oliveira, L.S., Petitjean, C., Heutte, L.: A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 63(7), 1455–1462 (2015)
Bayramoglu, N., Kannala, J., Heikkilä, J.: Deep learning for magnification independent breast cancer histopathology image classification. In: 2016 23rd International Conference on Pattern Recognition (ICPR), pp. 2440–2445 (2016)
Litjens, G., Bandi, P., Ehteshami Bejnordi, B., Geessink, O., Balkenhol, M., Bult, P., Halilovic, A., Hermsen, M., Loo, R., Vogels, R.: 1399 he-stained sentinel lymph node sections of breast cancer patients: the Camelyon dataset. GigaScience 7(6), 065 (2018)
Pati, P., Jaume, G., Foncubierta-Rodriguez, A., Feroce, F., Anniciello, A.M., Scognamiglio, G., Brancati, N., Fiche, M., Dubruc, E., Riccio, D.: Hierarchical graph representations in digital pathology. Med. Image Anal. 75, 102264 (2022)
Elmore, J.G., Longton, G.M., Carney, P.A., Geller, B.M., Onega, T., Tosteson, A.N., Nelson, H.D., Pepe, M.S., Allison, K.H., Schnitt, S.J.: Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313(11), 1122–1132 (2015)
Belsare, A., Mushrif, M., Pangarkar, M., Meshram, N.: Classification of breast cancer histopathology images using texture feature analysis. In: Tencon 2015–2015 IEEE Region 10 Conference, pp. 1–5 (2015)
Lei, B., Huang, S., Li, R., Bian, C., Li, H., Chou, Y.-H., Cheng, J.-Z.: Segmentation of breast anatomy for automated whole breast ultrasound images with boundary regularized convolutional encoder–decoder network. Neurocomputing 321, 178–186 (2018)
Hirsch, L., Huang, Y., Luo, S., Saccarelli, C.R., Gullo, R.L., Naranjo, I.D., Bitencourt, A.G., Onishi, N., Ko, E.S., Leithner, D.: Deep learning achieves radiologist-level performance of tumor segmentation in breast MRI (2020). arXiv preprint arXiv:2009.09827
Deniz, E., Şengür, A., Kadiroğlu, Z., Guo, Y., Bajaj, V., Budak, U.: Transfer learning based histopathologic image classification for breast cancer detection. Health Inf. Sci. Syst. 6, 1–7 (2018)
Yao, H., Zhang, X., Zhou, X., Liu, S.: Parallel structure deep neural network using CNN and RNN with an attention mechanism for breast cancer histology image classification. Cancers 11(12), 1901 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, pp. 234–241 (2015)
Lou, A., Guan, S., Loew, M.: Caranet: context axial reverse attention network for segmentation of small medical objects. J. Med. Imaging 10(1), 014005–014005 (2023)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition (2014). arXiv preprint arXiv:1409.1556
Fan, D.-P., Cheng, M.-M., Liu, Y., Li, T., Borji, A.: Structure-measure: a new way to evaluate foreground maps. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 4548–4557 (2017)
Qin, X., Zhang, Z., Huang, C., Dehghan, M., Zaiane, O.R., Jagersand, M.: U2-net: going deeper with nested u-structure for salient object detection. Pattern Recogn. 106, 107404 (2020)
Kim, T., Lee, H., Kim, D.: Uacanet: uncertainty augmented context attention for polyp segmentation. In: Proceedings of the 29th ACM International Conference on Multimedia, pp. 2167–2175 (2021)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition (2014). arXiv preprint arXiv:1409.1556
Funding
This work was funded by the National Natural Science Foundation of China (Grant No.32100530), Natural Science Foundation of Chongqing, China (Grant No. CSTB2022BSXM-JCX0065), the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN202200455). This work was also supported by Graduate Research and Innovation Fund of Yunnan University under Grants KC-22221913.
Author information
Authors and Affiliations
Contributions
Junjie Li and Kaixiang Yan wrote the main manuscript text. Yu Yu and Junjie Li prepared data. Lingyu Li provided primary ideal. Lingyu Li and Xiaohui Zhan proofread the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
To the best of our knowledge, the named authors have no conflict of interest, financial, or otherwise.
Ethical approval
No human or animal experiments are involved in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junjie Li and Kaixiang Yan contributted equally for this work.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Yan, K., Yu, Y. et al. BIS5k: a large-scale dataset for medical segmentation task based on HE-staining images of breast cancer. SIViP 18, 3705–3713 (2024). https://doi.org/10.1007/s11760-024-03034-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11760-024-03034-2