Abstract
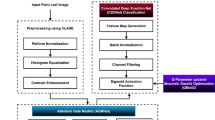
Agriculture faces considerable challenges with the growth of the demand for food. Potatoes are considered a staple food in many nations. The crop yield is affected by fungal pathogen diseases like Early Blight (EB) and Late Blight (LB). The crop yield can be enhanced by detecting the fungal pathogens at early stages using computer vision and artificial intelligence. The potato leaf disease segmentation is challenging in detecting the EB and LB. This paper presents a novel UNet-based architecture ARM-UNet (Attention-gate Residual path Modified UNet) in detecting potato fungal pathogen diseases. The proposed model replaces the skip connection by integrating the attention gates and residual paths to improve saliency and increase the depth of the network layers, which enhances the corresponding extracted pixel information transformed between the encoder and the decoder path. The ARM-UNet model segmented the diseased potato leaves. The proposed model performance is compared with the state-of-the-art UNet models and the existing methods for segmenting the diseased leaves using the performance metrics dice-score and mean-intersection over Union. The proposed model has produced an accuracy of 97.65%, where the DS is 93.15% and the mean-IoU is 84.18%. The experimental results show that the proposed model has efficiently segmented the diseased potato leaves.









Similar content being viewed by others

Data availability
No datasets were generated or analysed during the current study.
References
FAO: The Future of Food and Agriculture. Food and Agriculture Organization of the United Nations. 1–52: (2017)
FAO: World food and agriculture—statistical yearbook 2020: (2020)
Savary, S., Willocquet, L., Pethybridge, S.J., Esker, P., McRoberts, N., Nelson, A.: The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439 (2019). https://doi.org/10.1038/s41559-018-0793-y
Lin, X., Olave-Achury, A., Heal, R., Pais, M., Witek, K., Ahn, H.K., Zhao, H., Bhanvadia, S., Karki, H.S., Song, T., Wu, C., hang, Adachi, H., Kamoun, S., Vleeshouwers, V.G.A.A., Jones, J.D.G.: A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol. Plant. 15, 1457–1469 (2022). https://doi.org/10.1016/j.molp.2022.07.012
Stridh, L.J., Mostafanezhad, H., Andersen, C.B., Odilbekov, F., Grenville-Briggs, L., Lankinen, Å., Liljeroth, E.: Reduced efficacy of biocontrol agents and plant resistance inducers against potato early blight from greenhouse to field. J. Plant Dis. Prot. 129, 923–938 (2022). https://doi.org/10.1007/s41348-022-00633-4
Akino, S., Takemoto, D., Hosaka, K.: Phytophthora infestans: A review of past and current studies on potato late blight. J. Gen. Plant Pathol. 80, 24–37 (2014)
Zhu, T., Li, J., Chen, J., Li, J., Shu, G., Liu, R., Wu, J., Yang, Y., Liu, X., Zhou, X.: A rapid method for removing complex backgrounds for potato early blight monitoring. In: 2023 IEEE 9th International conference on cloud computing and intelligent systems (CCIS). pp. 568–572. IEEE (2023)
Anam, S., Fitriah, Z.: Early blight disease segmentation on tomato plant using K-means algorithm with swarm intelligence-based algorithm. Int. J. Math. Comput. Sci. 16, 1217–1228 (2021)
Sengar, N., Dutta, M.K., Travieso, C.M.: Computer vision based technique for identification and quantification of powdery mildew disease in cherry leaves. Computing. 100, 1189–1201 (2018)
Wang, Z., Wang, K., Yang, F., Pan, S., Han, Y.: Image segmentation of overlapping leaves based on Chan–Vese model and Sobel operator. Inform. Process. Agric. 5, 1–10 (2018)
Smith, A.G., Petersen, J., Selvan, R., Rasmussen, C.R.: Segmentation of roots in soil with U-Net. Plant. Methods. 16, 1–15 (2020)
Lin, K., Gong, L., Huang, Y., Liu, C., Pan, J.: Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front. Plant. Sci. 10, 155 (2019)
Srilakshmi, A., Geetha, K.: A novel framework for soybean leaves disease detection using DIM-U-net and LSTM. Multimed Tools Appl. 1–21 (2023)
Abinaya, S., Kumar, K.U., Alphonse, A.S.: Cascading autoencoder with attention residual U-Net for multi-class plant leaf disease segmentation and classification. IEEE Access. (2023)
Zou, K., Chen, X., Wang, Y., Zhang, C., Zhang, F.: A modified U-Net with a specific data argumentation method for semantic segmentation of weed images in the field. Comput. Electron. Agric. 187, 106242 (2021)
Tassis, L.M., de Souza, J.E.T., Krohling, R.A.: A deep learning approach combining instance and semantic segmentation to identify diseases and pests of coffee leaves from in-field images. Comput. Electron. Agric. 186, 106191 (2021)
Zhang, T., Yang, Z., Xu, Z., Li, J.: Wheat yellow rust severity detection by efficient DF-UNet and UAV multispectral imagery. IEEE Sens. J. 22, 9057–9068 (2022)
Fu, J., Zhao, Y., Wu, G.: Potato Leaf Disease Segmentation Method based on Improved UNet. Appl. Sci. 13, 11179 (2023)
Lee, S.-E., Kim, J.-O.: Multi-scale attention based plant disease segmentation network. In: 2023 International Technical conference on circuits/systems, computers, and communications (ITC-CSCC). pp. 1–4. IEEE (2023)
Deng, Y., Xi, H., Zhou, G., Chen, A., Wang, Y., Li, L., Hu, Y.: An effective image-based tomato leaf disease segmentation method using MC-UNet. Plant. Phenomics. 5, 0049 (2023)
Alex Lavaee: PlantifyDr Dataset | Kaggle, https://www.kaggle.com/lavaman151/plantifydr-dataset
Rifai, A.M., Raharjo, S., Utami, E., Ariatmanto, D.: Analysis for diagnosis of pneumonia symptoms using chest X-ray based on MobileNetV2 models with image enhancement using white balance and contrast limited adaptive histogram equalization (CLAHE). Biomed. Signal. Process. Control. 90, 105857 (2024)
Guoqiang, W., Hongxia, Z., Zhiwei, G., Wei, S., Dagong, J.: Bilateral filter denoising of Lidar point cloud data in automatic driving scene. Infrared Phys. Technol. 131, 104724 (2023)
Bougourzi, F., Distante, C., Dornaika, F., Taleb-Ahmed, A.: PDAtt-Unet: Pyramid dual-decoder attention Unet for Covid-19 infection segmentation from CT-scans. Med. Image Anal. 86, 102797 (2023)
Chinnam, S.K.R., Sistla, V., Kolli, V.K.K.: Multimodal attention-gated cascaded U-Net model for automatic brain tumor detection and segmentation. Biomed. Signal. Process. Control. 78, 103907 (2022)
Al-Masni, M.A., Al-Antari, M.A., Choi, M.-T., Han, S.-M., Kim, T.-S.: Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 162, 221–231 (2018)
Oktay, O., Schlemper, J., Folgoc, L., Le, Lee, M., Heinrich, M., Misawa, K., Mori, K., McDonagh, S., Hammerla, N.Y., Kainz, B.: Attention u-net: Learning where to look for the pancreas. (2018). arXiv preprint arXiv:1804.03999
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: A nested u-net architecture for medical image segmentation. In: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018. pp. 3–11 (2018)
Author information
Authors and Affiliations
Contributions
Author contributions D N Kiran Pandiri proposed the main idea and designed the main aspects of the project; and finalized the paper writing, reviewed, and edited it.R Murugan designed and implemented all the software required to perform the experiments and reviewed, and edited it.Tripti Goel evaluated the data and reviewed and edited it.
Corresponding author
Ethics declarations
Ethical approval
Ethical approval was not sought for the present study because the datasets utilized in this study are publicly available.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pandiri, D.N.K., Murugan, R. & Goel, T. ARM-UNet: attention residual path modified UNet model to segment the fungal pathogen diseases in potato leaves. SIViP 19, 80 (2025). https://doi.org/10.1007/s11760-024-03566-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11760-024-03566-7