Abstract
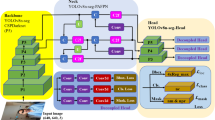
To monitor the condition of cupping spots in real-time during the operation of the automatic cupping machine, reduce the influence of the surrounding environment on the image, and improve the segmentation accuracy of the cupping spots, this paper proposes a network called MCA-Deeplabv3+. Firstly, backbone network replaced by Mobilenetv2 to reduce the model size and improve feature extraction speed; Secondly, to further enhance the network’s feature extraction capabilities, we added dilated convolution channels and integrated the CA attention mechanism into the ASPP module; Finally, data augmentation and brightness adjustment are performed on the dataset to improve the generalization of the model in different environments. The experimental results show that, in comparison with other segmentation models, MCA-Deeplabv3+performs the best in cupping spot segmentation, with mIoU and mPA reaching 93.90% and 96.73%, respectively. The practicality and effectiveness of the cupping spot segmentation model presented in this paper are thoroughly demonstrated.










Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Chirali, I.Z.: Cosmetic Cupping Therapy. In: Chirali, I.Z. (ed.) Traditional Chinese Medicine Cupping Therapy, 3rd edn., pp. 123–143. Elsevier, Amsterdam (2014)
Liu, Y.B., Zeng, Y.H., Qin, J.H.: GSC-YOLO: a lightweight network for cup and piston head detection. Signal, Image Video Process. 18(1), 351–360 (2024)
Sohail, A., Nawaz, N.A., Shah, A.A., Rasheed, S., Ilyas, S., Ehsan, M.K.: A systematic literature review on machine learning and deep learning methods for semantic segmentation. IEEE Access 10, 134557–134570 (2022). https://doi.org/10.1109/ACCESS.2022.3230983
Minaee, S., Boykov, Y., Porikli, F., et al.: Image segmentation using deep learning: a survey. IEEE Trans. Pattern Anal. Mach. Intell. 44(7), 3523–3542 (2021)
Wang, R., Lei, T., Cui, R., Zhang, B., Meng, H., Nandi, A.K.: Medical image segmentation using deep learning: a survey. IET Image Process. 16(5), 1243–1267 (2022)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 2015 pp. 3431–3440.
Ronneberger, O., Fischer, P., and Brox, T.: "U-Net: convolutional networks for biomedical image segmentation", Proc. 18th Int. Conf. Med. Image Comput. Comput.-Assist. Intervent., pp. 234–241, 2015.
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N., & Liang, J. (2018). Unet++: A nested u-net architecture for medical image segmentation. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Proceedings 4, 2018, Granada, Spain (pp. 3–11). https://doi.org/10.1007/978-3-030-00889-5_1.
Li, X., Fu, C., Wang, Q., Zhang, W., Sham, C.W., Chen, J.: DMSA-UNet: dual multi-scale attention makes UNet more strong for medical image segmentation. Knowl. Based Syst. (2024). https://doi.org/10.1016/j.knosys.2024.112050
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., Adam, H.: Encoder–decoder with Atrous separable convolution for semantic image segmentation, In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 2018 pp. 833–851.
Xiang, S., Wei, L., Hu, K.: Lightweight colon polyp segmentation algorithm based on improved DeepLabV3+. J. Cancer 15(1), 41–53 (2024). https://doi.org/10.7150/jca.88684
Tang, Y., Tan, D., Li, H., et al.: RTC_TongueNet: an improved tongue image segmentation model based on DeepLabV3. Digit. Health. (2024). https://doi.org/10.1177/20552076241242773
Liu, Y., Bai, X., Wang, J., Li, G., Li, J., Lv, Z.: Image semantic segmentation approach based on DeepLabV3 plus network with an attention mechanism. Eng. Appl. Artif. Intell. 1(127), 107260 (2024)
Xie, Z., Lu, Q., Guo, J., Lin, W., Ge, G., Tang, Y., Pasini, D., Wang, W.: Semantic segmentation for tooth cracks using improved DeepLabv3+ model. Heliyon. Feb 10(4) 2024.
Zhang, X., Bian, H., Cai, Y., et al.: An improved tongue image segmentation algorithm based on Deeplabv3+ framework. IET Image Proc. 16, 1473–1485 (2022)
Chollet, F.: Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) pp. 1800–1807.
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A. & Chen, L.-C. MobileNetV2: inverted residuals and linear bottlenecks. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 4510–4520. https://doi.org/10.1109/cvpr.2018.00474(2018).
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., \ Weyand, T. et al.: "MobileNets: efficient convolutional neural networks for mobile vision applications", arXiv:1704.04861, 2017.
Quach, L.-D., Quoc, K.N., Quynh, A.N., Ngoc, H.T., Thai-Nghe, N.: Tomato health monitoring system: tomato classification, detection, and counting system based on YOLOv8 model with explainable mobilenet models using grad-CAM++. IEEE Access 12, 9719–9737 (2024). https://doi.org/10.1109/ACCESS.2024.3351805
Li, X., Du, J., Yang, J., Li, S.: When mobilenetv2 meets transformer: a balanced sheep face recognition model. Agriculture 12(8), 1126 (2022). https://doi.org/10.3390/agriculture12081126
Chen, C., Li, B.: A Transform module to enhance lightweight attention by expanding receptive field. Expert Syst. Appl. 15(248), 123359 (2024). https://doi.org/10.1016/j.eswa.2024.123359
Hou, Q., Zhou, D., Feng, J.: Coordinate attention for efficient mobile network design. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 13713–13722. (2021)
Ming, Q., Xiao, X.: Towards accurate medical image segmentation with gradient-optimized dice loss. IEEE Signal Process. Lett. 31, 191–195 (2024). https://doi.org/10.1109/LSP.2023.3329437
Xie, Y., Ling, J.: Wood defect classification based on lightweight convolutional neural networks. BioResources 18(4), 7663–7680 (2023)
Funding
This work was supported by the National Natural Science Foundation of China (No. 61961011) and Guangxi Natural Science Foundation (No. 2021GXNSFAA220091).
Author information
Authors and Affiliations
Contributions
L-YM contributed to the development of the methodology, the investigation, the formal analysis, and the writing of the original draft. J-HQ contributed to the conceptualization, methodology, and visualization. Y-BL and T-TH contributed to the investigation, formal analysis, and writing review and editing. G-FZ and B-LX assisted with project administration and supervision. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
We have obtained written informed consent from all study participants.
Ethics approval and consent to participate
The study protocol was approved by the ethics review board of Guilin University of Technology. All of the procedures were performed in accordance with the Declaration of Helsinki and relevant policies in China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, LY., Qin, JH., Liu, YB. et al. MCA-Deeplabv3+: a cupping spot image segmentation network based on improved Deeplabv3+. SIViP 19, 187 (2025). https://doi.org/10.1007/s11760-024-03781-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11760-024-03781-2