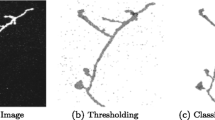
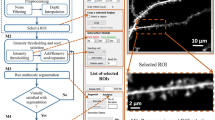
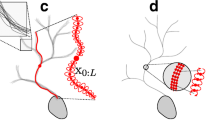
Abstract
The challenges faced in analyzing optical imaging data from neurons include a low signal-to-noise ratio of the acquired images and the multiscale nature of the tubular structures that range in size from hundreds of microns to hundreds of nanometers. In this paper, we address these challenges and present a computational framework for an automatic, three-dimensional (3D) morphological reconstruction of live nerve cells. The key aspects of this approach are: (i) detection of neuronal dendrites through learning 3D tubular models, and (ii) skeletonization by a new algorithm using a morphology-guided deformable model for extracting the dendritic centerline. To represent the neuron morphology, we introduce a novel representation, the Minimum Shape-Cost (MSC) Tree that approximates the dendrite centerline with sub-voxel accuracy and demonstrate the uniqueness of such a shape representation as well as its computational efficiency. We present extensive quantitative and qualitative results that demonstrate the accuracy and robustness of our method.
























Similar content being viewed by others
Notes
Loops are not allowed in a tree structure.
The coordinates of the points in the soma-pipette region are used to obtain the center of the ellipse given by the mean of the points and the semi-axis of the ellipse given by the covariance matrix.
The solution of Eq. (4) coincides with the centerline of each dendritic branch since small distances of a voxel from V D are penalized and the only points that are equidistant from the dendritic boundary are those of the centerline.
References
Bas, E., & Erdogmus, D. (2011). Principal curves as skeletons of tubular objects. Neuroinformatics, 9(2–3), 181–191.
Basu, S., Ooi, W.T., Racoceanu, D. (2014). Improved marked point process priors for single neurite tracing. In IEEE international workshop on Pattern recognition in neuroimaging (pp. 1–4).
Breitenreicher, D., Sofka, M., Britzen, S., Zhou, S. (2013). Hierarchical discriminative framework for detecting tubular structures in 3D images. In J. Gee (Ed.), Proceedings of information processing in medical imaging,Lecture Notes in computer Science (Vol. 7917, pp. 328–339). Asilomar, CA: Springer-Verlag Berlin Heidelberg.
Broser, P., Schulte, R., Roth, A., Helmchen, F., Lang, S., Wittum, G., Sakmann, B. (2004). Nonlinear anisotropic diffusion filtering of three-dimensional image data from two-photon microscopy. Journal of Biomedical Optics, 9(6), 1253–1264.
Brown, K., Barrionuevo, G., Canty, A., Paola, V., Hirsch, J., Jefferis, G., Lu, J., Snippe, M., Sugihara, I., Ascoli, G. (2011). The DIADEM data sets: representative light microscopy images of neuronal morphology to advance automation of digital reconstructions. Neuroinformatics, 9(2-3), 143–157.
Chothani, P., Mehta, V., Stepanyants, A. (2011). Automated tracing of neurites from light microscopy stacks of images. Neuroinformatics, 9(2–3), 263–278.
Computational Biomedicine Lab (2014). ORION: Online Reconstruction and functional Imaging Of Neurons. http://www.cbl.uh.edu/ORION.
Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning, 20(3), 273–297.
Cuntz, H., Forstner, F., Borst, A., Häusser, M., Morrison, A. (2010). One rule to grow them all: A general theory of neuronal branching and its practical application. PLoS Computational Biology, 6(8).
Diestel, R. (2005). Graph theory. Berlin Heidelberg New York: Springer.
Dijkstra, E. (1959). A note on two problems in connection with graphs. Numerische Mathematic, 1, 269–271.
Dima, A., Scholz, M., Obermayer, K. (2002). Automatic segmentation and skeletonization of neurons from confocal microscopy images based on the 3-D wavelet transform. IEEE Transactions on Image Processing, 11(7), 790–801.
Duke/Southampton (2013). Neuroscience research group, school of biological sciences, southampton university: Duke/southampton archive of neuronal morphology. http://neuron.duke.edu/cells/.
Evers, J., Schmitt, S., Sibila, M., Duch, C. (2005). Progress in functional neuroanatomy: Precise automatic geometric reconstruction of neuronal morphology from confocal image stacks. Journal of Neurophysiology, 93(4), 2331–2342.
Frangi, A., Niessen, W., Vincken, K., Viergever, M. (1998). Multiscale vessel enhancement filtering. In Proceedings of medical image computing and computer assisted intervention, (Vol. 1496 pp. 130–137). Cambridge, MA.
Gillette, T., Brown, K., Ascoli, A. (2011). The DIADEM metric: comparing multiple reconstructions of the same neuron. Neuroinformatics, 9(2–3), 233–245.
Gonzalez, G., Fleuret, F., Fua, P. (2009). Learning rotational features for filament detection. In Proceedings of IEEE computer society conference on computer vision and pattern recognition (pp. 1582–1589). Miami Beach, FL.
González, G., Turetken, E., Fleuret, F., Fua, P. (2010). Delineating trees in noisy 2D images and 3D image-stacks. In Proceedings of IEEE computer society conference on computer vision and pattern recognition (pp. 2799–2806). San Francisco, CA.
Greenspan, H., Laifenfeld, M., Einav, S., Barnea, O. (2001). Evaluation of center-line extraction algorithms in quantitative coronary angiography. IEEE Transactions on Medical Imaging, 20(9), 928–941.
Hassouna, M., Farag, A., Falk, R. (2005). Differential Fly-Throughs (DFT): A general framework for computing flight paths. In Proceedings of medical image computing and computer-assisted intervention, (Vol. 1 pp. 654–661). PalmSprings, CA
Kakadiaris, I., Santamaría-Pang, A., Colbert, C., Saggau, P. (2008). Automatic 3-D morphological reconstruction of neuron cells from multiphoton images. In J. Rittscher, R. Machiraju, S. Wong (Eds.), Microscopic image analysis for life science applications, (pp. 389–399). Norwood, MA: Artech House.
Lorenz, C., Carlsen, I., Buzug, T., Fassnacht, C., Weese, J. (1997). Multi-scale line segmentation with automatic estimation of width, contrast and tangential direction in 2D and 3D medical images. In Proceedings of first joint conference on computer vision, virtual reality and robotics in medicine and medial robotics and computer-assisted surgery, (Vol. 1205 pp. 233–244).
Losavio, B., Liang, Y., Santamaría-Pang, A., Kakadiaris, I., Colbert, C., Saggau, P. (2008). Live neuron morphology automatically reconstructed from multiphoton and confocal imaging data . Journal of Neurophysiology, 100, 2422–2429.
Mayerich, D., Bjornsson, C., Taylor, J., Roysam, B. (2012). NetMets: software for quantifying and visualizing errors in biological network segmentation. BMC bioinformatics, 13 (Suppl 8), S7.
Meijering, E. (2010). Neuron tracing in perspective. Cytometry Part A, 77A(7), 693–704.
Ming, X., Li, A., Wu, J., Yan, C., Ding, W., Gong, H., Zeng, S., Liu, Q. (2013). Rapid reconstruction of 3D neuronal morphology from light microscopy images with augmented rayburst sampling. PloS one, 8(12), e84,557.
Mizrahi, A., Ben-Ner, E., Katz, M., Kedem, K., Glusman, J., Libersat, F. (2000). Comparative analysis of dendritic architecture of identified neurons using the Hausdorff distance metric. The Journal of Comparative Neurology, 233(3), 415–428.
Neurolucida (2014). MBF Bioscience: stereology and neuron morphology quantitative analysis. http://www.mbfbioscience.com.
Pawley, J. (2006). Handbook of Biological Confocal Microscopy. New York: Springer-Verlag.
Pelt, J.v., & Schierwagen, A. (2004). Morphological analysis and modeling of neuronal dendrites. Mathematical Biosciences, 188(1-2), 147–155.
Peng, H., Long, F., Myers, G. (2011). Automatic 3D neuron tracing using all-path pruning. Bioinformatics, 27(13), i239.
Perona, P., & Malik, J. (1990). Scale-space and edge detection using anisotropic diffusion. IEEE Transactions on Pattern Analysis and Machine Intelligence, 12(7), 629–639.
Platt, J.C. (2000). Probabilistic outputs for support vector machines and comparison to regularize likelihood methods. Advances in large margin classifiers, (pp. 61–74).
Rall, W. (1977). Handbook of Physiology: The Nervous System. Chapter Core conductor theory and cable properties of neurons (Vol. 1, pp. 39–98). Baltimore, MD: American Physiological Society Bethesda.
Rosenfeld, A., & Pfaltz, J. (1968). Distance functions on digital pictures. Pattern Recognition, 1(1), 33–61.
Rouchdy, Y., & Cohen, L. (2009). The shading zone problem in geodesic voting and its solutions for the segmentation of tree structures. application to the segmentation of microglia extensions. In Proceedings of computer vision and pattern recognition workshops (pp. 66–71). Miami
Santamaría-Pang, A., Bildea, T., Colbert, C., Saggau, P., Kakadiaris, I. (2006). Towards segmentation of irregular tubular structures in 3D confocal microscope images. In Proceedings of miccai workshop in microscopic image analysis and applications in biology (pp. 78–85). Denmark, Copenhagen.
Santamaría-Pang, A., Colbert, C., Losavio, B., Saggau, P., Kakadiaris, I. (2007a). Automatic morphological reconstruction of neurons from optical images. In Proceedings of international workshop in microscopic image analysis and applications in biology. Piscataway.
Santamaría-Pang, A., Colbert, C., Saggau, P., Kakadiaris, I. (2007b). Automatic centerline extraction of irregular tubular structures using probability volumes from multiphoton imaging. In Proceedings of medical image computing and computer-assisted intervention (pp. 486–494). Brisbane
Santamaría-Pang, A., Bildea, T., Tan, S., Kakadiaris, I. (2008). Denoising for 3-D photon-limited imaging data using nonseparable filterbanks. IEEE Transactions on Image Processing, 17(12), 2312–2323.
Sato, Y., Nakajima, S., Atsumi, H., Koller, T., Gerig, G., Yoshida, S., Kikinis, R. (1998). 3-D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Medical Image Analysis, 2(2), 143–168.
Sethian, J. (1996). Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision and Materials Sciences. Cambridge: Cambridge University Press.
Sholl, D. (1953). Dendritic organization in the neurons of the visual and motor cortices of the cat. Journal of Anatomy, 87(4), 387–406.
Srinivasan, R., Li, Q., Zhou, X., Lu, J., Lichtman, J., Wong, S. (2010). Reconstruction of the neuromuscular junction connectome. Bioinformatics, 26(12), i64–i70.
Turetken, E., Benmansour, F., Fua, P. (2012). Automated reconstruction of tree structures using path classifiers and mixed integer programming. In Proceedings of IEEE conference on computer vision and pattern recognition (CVPR) (pp. 566–573). Rhode Island.
Turetken, E., Benmansour, F., Andres, B., Pfister, H., Fua, P. (2013). Reconstructing loopy curvilinear structures using integer programming. In Proceedings of the IEEE, CVPR (pp. 1822–1829). Portland.
Uehara, C., Colbert, C.M., Saggau, P., Kakadiaris, I. (2004). Towards automatic reconstruction of dendrite morphology from live neurons. In Proceedings of 26th annual international conference of the IEEE engineering in medicine and biology society (pp. 1798–1801). San Fransisco.
Urban, S., O’Malley, S.M., Walsh, B., Santamaria-Pang, A., Saggau, P., Colbert, C., Kakadiaris, I. (2006). Automatic reconstruction of dendrite morphology from optical section stacks. In Proceedings of 2nd international workshop on computer vision approaches to medical image analysis (pp. 190–201). Graz: Springer.
Uylings, H., & van Pelt, J. (2002). Measures for quantifying dendritic arborizations. Network: Computation in Neural Systems, 13(3), 397–414.
Vapnik, V. (1995). The Nature of Statistical Learning Theory. Berlin: Springer-Verlag.
Vasilkoski, Z., & Stepanyants, A. (2009). Detection of the optimal neuron traces in confocal microscopy images. Journal of Neuroscience Methods, 178(1), 197–204.
Wang, Y., Narayanaswamy, A., Roysam, B. (2011a). Novel 4-D open-curve active contour and curve completion approach for automated tree structure extraction. In Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE (pp. 1105–1112). Colorado Springs.
Wang, Y., Narayanaswamy, A., Tsai, C.-L., Roysam, B. (2011b). A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinformatics, 9(2-3), 193–217.
Wearne, S., Rodriguez, A., Ehlenberger, D., Rocher, A., Henderson, S., Hof, P. (2005). New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience, 136(3), 661–680.
Xiao, H., & Peng, H. (2013). APP2: automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics, 29(11), 1448– 1454.
Xie, J., Zhao, T., Lee, T., Myers, E., Peng, H. (2010). Automatic neuron tracing in volumetric microscopy images with anisotropic path Searching. In Proceedings of the medical image computing and computer-assisted intervention (pp. 472–479). Beijing.
Xie, J., Zhao, T., Lee, T., Myers, E., Peng, H. (2011). Anisotropic path searching for automatic neuron reconstruction. Medical image analysis, 15(5), 680–689.
Yuan, X., Trachtenberg, J., Potter, S., Roysam, B. (2009). MDL constrained 3-D grayscale skeletonization algorithm for automated extraction of dendrites and spines from fluorescence confocal images. Neuroinformatics, 7, 213–232.
Zhao, T., Xie, J., Amat, F., Clack, F., Ahammad, P., Peng, H., Long, F., Myers, E. (2011). Automated reconstruction of neuronal morphology based on local geometrical and global structural models. Neuroinformatics, 9(2-3), 247–261.
Zhao, Y., Xiong, H., Zhang, K., Zhou, X. (2009). Equilibrium modeling for 3D curvilinear structure tracking of confocal microscopy images. In Proceedings of the IEEE international conference on image processing (pp. 2533–2536). Cairo.
Acknowledgments
We wish to thank all of the members of the ORION team (Computational Biomedicine Lab 2014) and especially Costa M. Colbert, Yong Liang, and Bradley E. Losavio. The data were acquired at P. Saggau’s Laboratory in the Department of Neuroscience of the Baylor College of Medicine. This work was supported in part by NIH 5R01EB001048-02, NSF-DMS 0915242, NHARP 003652-0136-2009 and the University of Houston–Eckhard Pfeiffer Endowment Fund. Any opinions, findings, conclusions or recommendations expressed in this material are those of the authors and may not reflect the views of UH, NIH, NHARP, or NSF.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Santamaría-Pang, A., Hernandez-Herrera, P., Papadakis, M. et al. Automatic Morphological Reconstruction of Neurons from Multiphoton and Confocal Microscopy Images Using 3D Tubular Models. Neuroinform 13, 297–320 (2015). https://doi.org/10.1007/s12021-014-9253-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-014-9253-2