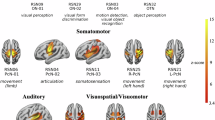
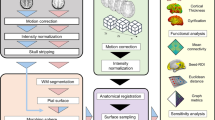
Abstract
Canonical resting state networks (RSNs) can be obtained through independent component analysis (ICA). RSNs are reproducible across subjects but also present inter-individual differences, which can be used to individualize regions-of-interest (ROI) definition, thus making fMRI analyses more accurate. Unfortunately, no automatic tool for defining subject-specific ROIs exists, making the classification of ICAs as representatives of RSN time-consuming and largely dependent on visual inspection. Here, we present Personode, a user-friendly and open source MATLAB-based toolbox that semi-automatically performs the classification of RSN and allows for defining subject- and group-specific ROIs. To validate the applicability of our new approach and to assess potential improvements compared to previous approaches, we applied Personode to both task-related activation and resting-state data. Our analyses show that for task-related activation analyses, subject-specific spherical ROIs defined with Personode produced higher activity contrasts compared to ROIs derived from single-study and meta-analytic coordinates. We also show that subject-specific irregular ROIs defined with Personode improved ROI-to-ROI functional connectivity analyses.
Hence, Personode might be a useful toolbox for ICA map classification into RSNs and group- as well as subject-specific ROI definitions, leading to improved analyses of task-related activation and functional connectivity.





Similar content being viewed by others
References
Beckmann, C. F., DeLuca, M., Devlin, J. T., & Smith, S. M. (2005). Investigations into resting-state connectivity using independent component analysis. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 360(1457), 1001–1013. https://doi.org/10.1098/rstb.2005.1634.
Bullmore, E. T., & Sporns, O. (2009). Complex brain networks: Graph theoretical analysis of structural and functional systems. Nature Reviews. Neuroscience, 10(3), 186–198. https://doi.org/10.1038/nrn2575.
Cameron Craddock, R., Holtzheimer, P. E., Hu, X. P., & Mayberg, H. S. (2009). Disease state prediction from resting state functional connectivity. Magnetic Resonance in Medicine, 62(6), 1619–1628. https://doi.org/10.1002/mrm.22159.
Dai, W., Varma, G., Scheidegger, R., & Alsop, D. C. (2016). Quantifying fluctuations of resting state networks using arterial spin labeling perfusion MRI. Journal of Cerebral Blood Flow & Metabolism, 36(3), 463–473. https://doi.org/10.1177/0271678X15615339.
Damoiseaux, J. S., Rombouts, S. A. R. B., Barkhof, F., Scheltens, P., Stam, C. J., Smith, S. M., & Beckmann, C. F. (2006). Consistent resting-state networks across healthy subjects. Proceedings of the National Academy of Sciences, 103(37), 13848–13853. https://doi.org/10.1073/pnas.0601417103.
Finn, E. S., Shen, X., Scheinost, D., Rosenberg, M. D., Huang, J., Chun, M. M., et al. (2015). Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nature Neuroscience, 18, 1664–1671. https://doi.org/10.1038/nn.4135.
Fox, M. D., Snyder, A. Z., Vincent, J. L., Corbetta, M., Van Essen, D. C., & Raichle, M. E. (2005). The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences, 102(27), 9673–9678. https://doi.org/10.1073/pnas.0504136102.
Golestani, A. M., & Goodyear, B. G. (2011). Regions of interest for resting-state fMRI analysis determined by inter-voxel cross-correlation. NeuroImage, 56(1), 246–251. https://doi.org/10.1016/j.neuroimage.2011.02.038.
Harrison, B. J., Pujol, J., López-Solà, M., Hernández-Ribas, R., Deus, J., Ortiz, H., Soriano-Mas, C., Yücel, M., Pantelis, C., & Cardoner, N. (2008). Consistency and functional specialization in the default mode brain network. Proceedings of the National Academy of Sciences, 105(28), 9781–9786. https://doi.org/10.1073/pnas.0711791105.
Liu, T. (2011). A few thoughts on brain ROIs. Brain Imaging and Behavior, 5(3), 189–202. https://doi.org/10.1007/s11682-011-9123-6.
De Luca, M., Beckmann, C. F., De Stefano, N., Matthews, P. M., & Smith, S. M. (2006). fMRI resting state networks define distinct modes of long-distance interactions in the human brain. NeuroImage, 29(4), 1359–1367. https://doi.org/10.1016/j.neuroimage.2005.08.035.
Mcdonald, A. R., Muraskin, J., Van Dam, N. T., Froehlich, C., Puccio, B., Pellman, J., et al. (2017). The real-time fMRI neurofeedback based stratification of default network regulation neuroimaging data repository. NeuroImage, 146, 157–170. https://doi.org/10.1016/j.neuroimage.2016.10.048.
Miller, K. L., Alfaro-Almagro, F., Bangerter, N. K., Thomas, D. L., Yacoub, E., Xu, J., Bartsch, A. J., Jbabdi, S., Sotiropoulos, S. N., Andersson, J. L., Griffanti, L., Douaud, G., Okell, T. W., Weale, P., Dragonu, I., Garratt, S., Hudson, S., Collins, R., Jenkinson, M., Matthews, P. M., & Smith, S. M. (2016). Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nature Neuroscience, 19(11), 1523–1536. https://doi.org/10.1038/nn.4393.
Nucifora, P. G. P., Verma, R., Lee, S.-K., & Melhem, E. R. (2007). Diffusion-tensor MR imaging and tractography: Exploring brain microstructure and connectivity. Radiology, 245(2), 367–384. https://doi.org/10.1148/radiol.2452060445.
Poldrack, R. A. (2007). Region of interest analysis for fMRI. Social Cognitive and Affective Neuroscience, 2(1), 67–70. https://doi.org/10.1093/scan/nsm006.
Rubinov, M., & Sporns, O. (2010). Complex network measures of brain connectivity: Uses and interpretations. NeuroImage, 52(3), 1059–1069. https://doi.org/10.1016/J.NEUROIMAGE.2009.10.003.
Schilbach, L., Bzdok, D., Timmermans, B., Fox, P. T., Laird, A. R., Vogeley, K., & Eickhoff, S. B. (2012). Introspective minds: Using ALE meta-analyses to study commonalities in the neural correlates of emotional processing, social & unconstrained cognition. PLoS One, 7(2), 1–10. https://doi.org/10.1371/journal.pone.0030920.
Seeley, W. W., Menon, V., Schatzberg, A. F., Keller, J., Glover, G. H., Kenna, H., Reiss, A. L., & Greicius, M. D. (2007). Dissociable intrinsic connectivity networks for salience processing and executive control. Journal of Neuroscience, 27(9), 2349–2356. https://doi.org/10.1523/JNEUROSCI.5587-06.2007.
Sitaram, R., Ros, T., Stoeckel, L., Haller, S., Scharnowski, F., Lewis-Peacock, J., et al. (2016). Closed-loop brain training: The science of neurofeedback. Nature Reviews Neuroscience, 18, 86.
Smith, S. M., Fox, P. T., Miller, K. L., Glahn, D. C., Fox, P. M., Mackay, C. E., Filippini, N., Watkins, K. E., Toro, R., Laird, A. R., & Beckmann, C. F. (2009). Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences, 106(31), 13040–13045. https://doi.org/10.1073/pnas.0905267106.
Sohn, W., Yoo, K., Lee, Y.-B., Seo, S., Na, D., & Jeong, Y. (2015). Influence of ROI selection on resting state functional connectivity: An individualized approach for resting state fMRI analysis. Frontiers in Neuroscience, 9, 280. https://doi.org/10.3389/fnins.2015.00280.
Sulzer, J., Haller, S., Scharnowski, F., Weiskopf, N., Birbaumer, N., Blefari, M. L., Bruehl, A. B., Cohen, L. G., DeCharms, R., Gassert, R., Goebel, R., Herwig, U., LaConte, S., Linden, D., Luft, A., Seifritz, E., & Sitaram, R. (2013). Real-time fMRI neurofeedback: Progress and challenges. NeuroImage, 76, 386–399. https://doi.org/10.1016/j.neuroimage.2013.03.033.
van den Heuvel, M. P., & Hulshoff Pol, H. E. (2010). Exploring the brain network: A review on resting-state fMRI functional connectivity. European Neuropsychopharmacology, 20(8), 519–534. https://doi.org/10.1016/j.euroneuro.2010.03.008.
Veer, I. M., Beckmann, C. F., van Tol, M.-J., Ferrarini, L., Milles, J., Veltman, D. J., et al. (2010). Whole brain resting-state analysis reveals decreased functional connectivity in major depression. Frontiers in Systems Neuroscience. https://doi.org/10.3389/fnsys.2010.00041.
Whitfield-Gabrieli, S., & Nieto-Castanon, A. (2012). Conn: A functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connectivity, 2(3), 125–141. https://doi.org/10.1089/brain.2012.0073.
Acknowledgements
This work was supported by the Brazilian National Council for Scientific and Technological Development (CNPq), the Brazilian National Council for the Improvement of Higher Education (CAPES), the Swiss National Science Foundation (BSSG10_155915, 100014_178841, 32003B_166566), the Foundation for Research in Science and the Humanities at the University of Zurich (STWF-17-012), the Baugarten Stiftung, and the Swiss Government. We also thank Dr. Ludovica Griffanti for insightful discussions and comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Information Sharing Statement
NKI/Rockland Sample (RRID:SCR_009435) data used in this work is available at https://fcon_1000.projects.nitrc.org/indi/enhanced/neurodata.html. UK Biobank (RRID:SCR_012815) group-averaged RSNs templates where obtained at http://biobank.ctsu.ox.ac.uk/showcase/refer.cgi?id=9028.
SPM12 (RRID:SCR_007037,https://www.fil.ion.ucl.ac.uk/spm/software/spm12/), MarsBaR (RRID:SCR_009605,http://marsbar.sourceforge.net), and GIFT (RRID:SCR_001953,http://trendscenter.org/trends/software/gift/) are available to the general public.
The source code of Personode is available free of charge for non-commercial use and adaptation, under the condition of proper attribution, at https://github.com/gustavopamplona/Personode.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(PDF 636 kb)
About this article
Cite this article
Pamplona, G.S.P., Vieira, B.H., Scharnowski, F. et al. Personode: A Toolbox for ICA Map Classification and Individualized ROI Definition. Neuroinform 18, 339–349 (2020). https://doi.org/10.1007/s12021-019-09449-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-019-09449-4