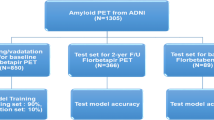
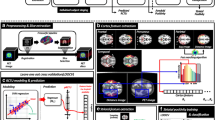
Abstract
Automated amyloid-PET image classification can support clinical assessment and increase diagnostic confidence. Three automated approaches using global cut-points derived from Receiver Operating Characteristic (ROC) analysis, machine learning (ML) algorithms with regional SUVr values, and deep learning (DL) network with 3D image input were compared under various conditions: number of training data, radiotracers, and cohorts. 276 [11C]PiB and 209 [18F]AV45 PET images from ADNI database and our local cohort were used. Global mean and maximum SUVr cut-points were derived using ROC analysis. 68 ML models were built using regional SUVr values and one DL network was trained with classifications of two visual assessments – manufacturer’s recommendations (gray-scale) and with visually guided reference region scaling (rainbow-scale). ML-based classification achieved similarly high accuracy as ROC classification, but had better convergence between training and unseen data, with a smaller number of training data. Naïve Bayes performed the best overall among the 68 ML algorithms. Classification with maximum SUVr cut-points yielded higher accuracy than with mean SUVr cut-points, particularly for cohorts showing more focal uptake. DL networks can support the classification of definite cases accurately but performed poorly for equivocal cases. Rainbow-scale standardized image intensity scaling and improved inter-rater agreement. Gray-scale detects focal accumulation better, thus classifying more amyloid-positive scans. All three approaches generally achieved higher accuracy when trained with rainbow-scale classification. ML yielded similarly high accuracy as ROC, but with better convergence between training and unseen data, and further work may lead to even more accurate ML methods.



Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Availability of Data and Material
The data will not be available as the authors have no permission to share the data.
References
Cattell, L., Platsch, G., Pfeiffer, R., Declerck, J., Schnabel, J. A., & Hutton, C. (2015). Classification of amyloid status using machine learning with histograms of oriented 3D gradients. NeuroImage Clinical, 12, 990–1003. https://doi.org/10.1016/j.nicl.2016.05.004
Eli Lilly. (2012). Highlights of prescribing information Amyvid (florbetapir F 18 injection). Revised December 2019 from https://pi.lilly.com/us/amyvid-uspi.pdf
Gibson, E., Li, W., Sudre, C., et al. (2018). NiftyNet: A deep-learning platform for medical imaging. Computer Methods and Programs in Biomedicine, 158, 113–122. https://doi.org/10.1016/j.cmpb.2018.01.025
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770–778). https://doi.org/10.1109/CVPR.2016.90
Hu, J., Shen, L., & Sun, G. (2018). Squeeze-and-Excitation Networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 7132–7141).
Jack, C. R., Bennett, D. A., Blennow, K., et al. (2018). NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s & Dementia, 14(4), 535–562. https://doi.org/10.1016/j.jalz.2018.02.018
Johnson, K. A., Minoshima, S., Bohnen, N. I., et al. (2013). Appropriate use criteria for Amyloid PET: A report of the Amyloid imaging task force the society of nuclear medicine and molecular imaging and the Alzheimer’s association. Journal of Nuclear Medicine, 54(3), 476–490. https://doi.org/10.2967/jnumed.113.120618
Kang, H., Kim, W. -G., Yang, G. -S., et al. (2018). VGG-based BAPL Score Classification of 18F-Florbetaben Amyloid Brain PET. Biomedical Science Letters, 24(4), 418–425. https://doi.org/10.15616/bsl.2018.24.4.418
Kim, J. P., Kim, J., Kim, Y., et al. (2020). Staging and quantification of florbetaben PET images using machine learning: Impact of predicted regional cortical tracer uptake and amyloid stage on clinical outcomes. European Journal of Nuclear Medicine and Molecular Imaging, 47(8), 1971–1983. https://doi.org/10.1007/s00259-019-04663-3
Krippendorff, K. (2011) Computing Krippendorff’s Alpha-Reliability (p. 12). Dep Pap. http://repository.upenn.edu/asc_papers
Lundeen, T. F., Seibyl, J. P., Covington, M. F., Eshghi, N., & Kuo, P. H. (2018). Signs and Artifacts in Amyloid PET. Radiographics, 38(7), 2123–2133. https://doi.org/10.1148/rg.2018180160
McHugh, M. L. (2012) Interrater reliability: The kappa statistic. Biochemia Medica, 22(3), 276–282. https://doi.org/10.11613/bm.2012.031
Ng, S., Villemagne, V. L., Berlangieri, S., et al. (2007). Visual assessment versus quantitative assessment of 11C-PIB PET and 18F-FDG PET for detection of Alzheimer’s disease. Journal of Nuclear Medicine, 48(4), 547–552. https://doi.org/10.2967/jnumed.106.037762
Reilhac, A., Merida, I., Irace, Z., et al. (2018). Development of a dedicated rebinner with rigid motion correction for the mMR PET/MR Scanner, and Validation in a Large Cohort of 11C-PIB Scans. Journal of Nuclear Medicine, 59(11), 1761–1767. https://doi.org/10.2967/jnumed.117.206375
Rowe, C. C., & Villemagne, V. L. (2013). Brain amyloid imaging. Journal of Nuclear Medicine Technology, 41(1), 11–18. https://doi.org/10.2967/jnumed.110.076315
Son, H. J., Oh, J. S., Oh, M., et al. (2020). The clinical feasibility of deep learning-based classification of amyloid PET images in visually equivocal cases. European Journal of Nuclear Medicine and Molecular Imaging, 47(2), 332–341. https://doi.org/10.1007/s00259-019-04595-y
Tanaka, T., Stephenson, M. C., Nai, Y. H., et al. (2020). Improved quantification of amyloid burden and associated biomarker cut-off points: Results from the first amyloid Singaporean cohort with overlapping cerebrovascular disease. European Journal of Nuclear Medicine and Molecular Imaging, 47(2), 319–331. https://doi.org/10.1007/s00259-019-04642-8
Vandenberghe, R., Nelissen, N., Salmon, E., et al. (2013). Binary classification of 18F-flutemetamol PET using machine learning: Comparison with visual reads and structural MRI. NeuroImage, 64(1), 517–525. https://doi.org/10.1016/j.neuroimage.2012.09.015
Yamane, T., Ishii, K., Sakata, M., et al. (2017). Inter-rater variability of visual interpretation and comparison with quantitative evaluation of 11C-PiB PET amyloid images of the Japanese Alzheimer’s Disease Neuroimaging Initiative (J-ADNI) multicenter study. European Journal of Nuclear Medicine and Molecular Imaging, 44(5), 850–857. https://doi.org/10.1007/s00259-016-3591-2
Acknowledgements
We acknowledge all the coordinators from NUH memory clinic and Memory Aging and Cognition Centre for their contributions in subject recruitment and data acquisition, and the PET radiochemistry team from CIRC for the production of [11C]PiB used in this study. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California
Funding
This study was supported by the following National Medical Research Council grant in Singapore: NMRC/CG/NUHS/2010-R-184–005-184–511, NMRC/CG/013/2013, and NMRC/CIRG/1446/2016.
Author information
Authors and Affiliations
Consortia
Contributions
Ying-Hwey Nai and Anthonin Reilhac designed and supervised the study. Yee-Hsin Tay, Tomotaka Tanaka, Anthonin Reilhac and Ying-Hwey Nai carried out the visual assessments. Yee-Hsin Tay and Ying-Hwey Nai and Anthonin Reilhac analyzed the data and wrote the draft manuscript. Edward G. Robins was responsible for the production and delivery of [11C]PiB used in the study. Christopher P. Chen designed the clinical study and is responsible for clinical data. All authors contributed to the revision of the draft manuscript and approved the final version of the manuscript for submission.
Corresponding author
Ethics declarations
Informed Consent
Informed consent was obtained from all individual participants included in the study. Patients signed informed consent regarding publishing their data and photographs.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Research Involving Human Participants
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Ethics approval was obtained from the National-Healthcare Group Domain-Specific Review Board in Singapore.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ying-Hwey Nai and Anthonin Reilhac made an equal contribution to this manuscript.
Alzheimer's Disease Neuroimaging Initiative (ADNI) is a Group/Institutional Author.
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nai, YH., Tay, YH., Tanaka, T. et al. Comparison of Three Automated Approaches for Classification of Amyloid-PET Images. Neuroinform 20, 1065–1075 (2022). https://doi.org/10.1007/s12021-022-09587-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-022-09587-2