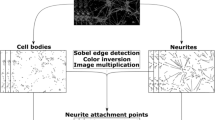
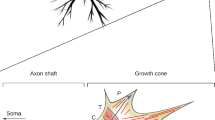
Abstract
Neuron morphology gives rise to distinct axons and dendrites and plays an essential role in neuronal functionality and circuit dynamics. In rat hippocampal neurons, morphological development occurs over roughly one week in vitro. This development has been qualitatively described as occurring in 5 stages. Still, there is a need to quantify cell growth to monitor cell culture health, understand cell responses to sensory cues, and compare experimental results and computational growth model predictions. To address this need, embryonic rat hippocampal neurons were observed in vitro over six days, and their processes were quantified using both standard morphometrics (degree, number of neurites, total length, and tortuosity) and new metrics (distance between change points, relative turning angle, and the number of change points) based on the Change-Point Test to track changes in path trajectories. Of the standard morphometrics, the total length of neurites per cell and the number of endpoints were significantly different between 0.5, 1.5, and 4 days in vitro, which are typically associated with Stages 2-4. Using the Change-Point Test, the number of change points and the average distance between change points per cell were also significantly different between those key time points. This work highlights key quantitative characteristics, both among common and novel morphometrics, that can describe neuron development in vitro and provides a foundation for analyzing directional changes in neurite growth for future studies.







Similar content being viewed by others
References
Bicknell, B. A., Pujic, Z., Dayan, P., & Goodhill, G. J. (2018). Control of neurite growth and guidance by an inhibitory cell-body signal. PLOS Computational Biology, 14, e1006218.
Boulan, B., Beghin, A., Ravanello, C., Deloulme, J.-C., Gory-Fauré, S., Andrieux, A., Brocard, J., & Denarier, E. (2020). AutoNeuriteJ: An ImageJ plugin for measurement and classification of neuritic extensions. PLOS ONE, 15, e0234529.
Byrne, R. W., Noser, R., Bates, L. A., & Jupp, P. E. (2009). How did they get here from there? Detecting changes of direction in terrestrial ranging. Animal Behaviour, 77, 619–631.
Conover, W. J. (1971). Practical Nonparametric Statistics. (1st ed.). John Wiley & Sons, Inc.
Cuntz, H., Borst, A., & Segev, I. (2007). Optimization principles of dendritic structure. Theoretical Biology and Medical Modelling, 4, 21.
Deinhardt, K., Kim, T., Spellman, D. S., Mains, R. E., Eipper, B. A., Neubert, T. A., Chao, M. V., & Hempstead, B. L. (2011). Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Science Signaling, 4, ra82.
Dinno, A. (2017). dunn.test: Dunn’s Test of Multiple Comparisons Using Rank Sums. R package version 1.3.5.
Dotti, C. G., Sullivan, C. A., & Banker, G. A. (1988). The establishment of polarity by hippocampal neurons in culture. Journal of Neuroscience, 8, 1454–1468.
Ferrante, M., Migliore, M., & Ascoli, G. A. (2013). Functional impact of dendritic branch-point morphology. Journal of Neuroscience, 33, 2156–2165.
Ferreira Castro, A., Baltruschat, L., Stürner, T., Bahrami, A., Jedlicka, P., Tavosanis, G., & Cuntz, H. (2020). Achieving functional neuronal dendrite structure through sequential stochastic growth and retraction. eLife, 9.
Gillette, T. A., & Grefenstette, J. J. (2009). On comparing neuronal morphologies with the constrained tree-edit-distance. Neuroinformatics, 7, 191–4.
Gross, J., & Ligges, U. (2015). nortest: Tests for Normality [Computer software manual]. Retrieved from https://CRAN.R-project.org/package=nortest. (R package version 1.0-4).
Heumann, H., & Wittum, G. (2009). The tree-edit-distance, a measure for quantifying neuronal morphology. Neuroinformatics, 7, 179–90.
Ho, S.-Y., Chao, C.-Y., Huang, H.-L., Chiu, T.-W., Charoenkwan, P., & Hwang, E. (2011). NeurphologyJ: An automatic neuronal morphology quantification method and its application in pharmacological discovery. BMC Bioinformatics, 12, 230.
Jefferis, G. S., Potter, C. J., Chan, A. M., Marin, E. C., Rohlfing, T., Maurer, C. R., & Luo, L. (2007). Comprehensive maps of Drosophila higher olfactory centers: Spatially segregated fruit and pheromone representation. Cell, 128, 1187–1203.
Kaech, S., & Banker, G. (2006). Culturing hippocampal neurons. Nature Protocols, 1, 2406–2415.
Kanari, L., Dłotko, P., Scolamiero, M., Levi, R., Shillcock, J., Hess, K., & Markram, H. (2018). A topological representation of branching neuronal morphologies. Neuroinformatics, 16, 3–13.
Kang, S., Chen, X., Gong, S., Yu, P., Yau, S., Su, Z., Zhou, L., Yu, J., Pan, G., & Shi, L. (2017). Characteristic analyses of a neural differentiation model from iPSC-derived neuron according to morphology, physiology, and global gene expression pattern. Scientific Reports, 7, 12233.
Khalil, R., Farhat, A., & Dłotko, P. (2021). Developmental changes in pyramidal cell morphology in multiple visual cortical areas using cluster analysis. Frontiers in Computational Neuroscience, 15, 667696.
Kim, K.-M., Son, K., & Palmore, G. T. R. (2015). Neuron image analyzer: Automated and accurate extraction of neuronal data from low quality images. Scientific Reports, 5, 17062.
Kluyver, T., Ragan-Kelley, B., Pérez, F., & Granger, B. (2016). Jupyter Notebooks - a publishing format for reproducible computational workflows. In F. Loizides & B. Schmidt (Eds.), Positioning and Power in Academic Publishing: Players, Agents and Agendas (pp. 87–90). IOS Press.
Krichmar, J. L., Nasuto, S. J., Scorcioni, R., Washington, S. D., & Ascoli, G. A. (2002). Effects of dendritic morphology on CA3 pyramidal cell electrophysiology: A simulation study. Brain Research, 941, 11–28.
Laturnus, S., Kobak, D., & Berens, P. (2020). A systematic evaluation of interneuron morphology representations for cell type discrimination. Neuroinformatics, 18, 591–609.
Li, A., Barati Farimani, A., & Zhang, Y. J. (2021). Deep learning of material transport in complex neurite networks. Scientific Reports, 11, 11280.
Li, A., Chai, X., Yang, G., & Zhang, Y. J. (2019). An isogeometric analysis computational platform for material transport simulation in complex neurite networks. Molecular & Cellular Biomechanics, 16, 123–140.
Li, A., & Zhang, Y. J. (2022a). Modeling intracellular transport and traffic jam in 3D neurons using PDE-constrained optimization. Journal of Mechanics, 38, 44–59.
Li, A., & Zhang, Y. J. (2022b). Modeling material transport regulation and traffic jam in neurons using PDE-constrained optimization. Scientific Reports, 12, 3902.
Liao, A. S., Webster-Wood, V. A., & Zhang, Y. J. (2021). Quantification of neuron morphological development using the change-point test. In 2021 Summer Biomechanics, Bioengineering and Biotransport Conference. Virtual.
Mainen, Z. F., & Sejnowski, T. J. (1996). Influence of dendritic structure on firing pattern in model neocortical neurons. Nature, 382, 363–366.
Meijering, E., Jacob, M., Sarria, J.-C., Steiner, P., Hirling, H., & Unser, M. (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry, 58A, 167–176.
Polavaram, S., Gillette, T. A., Parekh, R., & Ascoli, G. A. (2014). Statistical analysis and data mining of digital reconstructions of dendritic morphologies. Frontiers in Neuroanatomy, 8, 138.
Pool, M., Thiemann, J., Bar-Or, A., & Fournier, A. E. (2008). NeuriteTracer: A novel ImageJ plugin for automated quantification of neurite outgrowth. Journal of Neuroscience Methods, 168, 134–139.
Powell, S. K., Rivas, R. J., Rodriguez-Boulan, E., & Hatten, M. E. (1997). Development of polarity in cerebellar granule neurons. Journal of Neurobiology, 32, 223–236.
Python Core Team. (2021). Python: A Dynamic, Open Source Programming Language. Python Software Foundation. Retrieved from https://www.python.org/
Qian, K., Pawar, A., Liao, A., Anitescu, C., Webster-Wood, V., Feinberg, A., Rabczuk, T., & Zhang, Y. J. (2022). Modeling neuron growth using isogeometric collocation based phase field method. Scientific Reports, 12, 8120.
R Core Team. (2021). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org/
RStudio Team. (2021). RStudio: Integrated Development Environment for R. Boston, MA: RStudio, PBC. Retrieved from http://www.rstudio.com/
Rueden, C. T., Schindelin, J., Hiner, M. C., DeZonia, B. E., Walter, A. E., Arena, E. T., & Eliceiri, K. W. (2017). Image J2: ImageJ for the next generation of scientific image data. BMC Bioinformatics, 18, 529.
Schaefer, A. T., Larkum, M. E., Sakmann, B., & Roth, A. (2003). Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern. Journal of Neurophysiology, 89, 3143–3154.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. -Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P., & Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9, 676–682.
Sholl, D. A. (1953). Dendritic organization in the neurons of the visual and motor cortices of the cat. Journal of anatomy, 87, 387–406.
Su, C. -Z., Chou, K. -T., Huang, H. -P., Li, C. -J., Charng, C. -C., Lo, C. -C., & Wang, D. -W. (2021). Identification of neuronal polarity by node-based machine learning. Neuroinformatics, 19, 669–684.
Tahirovic, S., & Bradke, F. (2009). Neuronal polarity. Cold Spring Harbor Perspectives in Biology, 1, a001644–a001644.
Tamariz, E., & Varela-Echavarría, A. (2015). The discovery of the growth cone and its influence on the study of axon guidance. Frontiers in Neuroanatomy, 9, 51.
Thermo Fisher Scientific. (2018). B-27 Plus Neuronal Culture System. Life Technologies. Retrieved from https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFSAssets/LSG/manuals/MAN0017319_B27_PlusNeuronalCultureSystem_UG.pdf
Uylings, H. B. M., & van Pelt, J. (2002). Measures for quantifying dendritic arborizations. Network: Computation in Neural Systems, 13, 397–414.
van Elburg, R. A. J., & van Ooyen, A. (2010). Impact of dendritic size and dendritic topology on burst firing in pyramidal cells. PLoS Computational Biology, 6, e1000781.
Vetter, P., Roth, A., & Häusser, M. (2001). Propagation of action potentials in dendrites depends on dendritic morphology. Journal of Neurophysiology, 85, 926–937.
Waskom, M. (2021). Seaborn: Statistical data visualization. Journal of Open Source Software, 6, 3021.
Zomorrodi, R., Ferecskó, A. S., Kovács, K., Kröger, H., & Timofeev, I. (2010). Analysis of morphological features of thalamocortical neurons from the ventroposterolateral nucleus of the cat. The Journal of Comparative Neurology, 518, 3541–3556.
Acknowledgements
We thank the anonymous reviewers for helpful comments on an earlier version of this manuscript.
Funding
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE1745016, the Faculty Early Career Development Program under Grant No. ECCS-2044785 and the LEAP HI Program under Grant No. CMMI-1953323. The authors were also supported in part by a PITA (Pennsylvania Infrastructure Technology Alliance) grant and a PMFI (Pennsylvania Manufacturing Fellows Initiative) grant. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization: VAW, YJZ; Data Curation: (Lead) ASL, (Supporting) WC; Formal Analysis: ASL; Funding Acquisition: VAW, YJZ; Investigation: ASL; Methodology: ASL, VAW; Software: ASL; Supervision: VAW, YJZ; Visualization: ASL; Writing - Original Draft: ASL; Writing - Review & Editing: ASL, VAW, YJZ, (Supporting) WC
Corresponding author
Ethics declarations
Ethics Approval
Not Applicable.
Consent to Participate
Not Applicable.
Competing Interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix: Distributions and Analyses on All Morphometrics for All Observed Time Points
Appendix: Distributions and Analyses on All Morphometrics for All Observed Time Points
A summary of the Dunn tests along with each feature’s distributions are showcased in Fig. 8. The Dunn tests with a Bonferroni correction indicated significant differences between several time points, as outlined in the corresponding Tables below, for segment length (Fig. 8a, Table 10), number of change points (Fig. 8c, Table 12), total length (Fig. 8d, Table 13), number of neurites (Fig. 8e, Table 14), and degree (Fig. 8g, Table 16). No significant differences between time points were detected for turning angle (Fig. 8b, Table 11), and significant differences were only detected between DIV 1.5 and 3 for tortuosity (Fig. 8f, Table 15).
The distributions and results of the Dunn test with a Bonferroni correction used to assess each morphometric, a average segment length, b average relative turning angle, c number of change points; d total length, e number of neurites, f average tortuosity, g degree, for every time point pair are symbolically represented, as defined in h
Additionally, the sample sizes of the data set are reported in Table 1. The summary statistics and Anderson-Darling results for all of the morphometrics are detailed in Tables 2, 3, 4, 5, 6, 7 and 8. The \(\chi ^2\) and \(p\)-values from the Kruskal-Wallis tests for each feature are in Table 9. Lastly, the post-hoc Dunn tests with a Bonferroni correction \(p\)-values are in Tables 10, 11, 12, 13, 14, 15 and 16.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liao, A.S., Cui, W., Zhang, Y.J. et al. Semi-Automated Quantitative Evaluation of Neuron Developmental Morphology In Vitro Using the Change-Point Test. Neuroinform 21, 163–176 (2023). https://doi.org/10.1007/s12021-022-09600-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-022-09600-8