Abstract
The brain is composed of a dense and ramified vascular network of arteries, veins and capillaries of various sizes. One way to assess the risk of cerebrovascular pathologies is to use computational models to predict the physiological effects of reduced blood supply and correlate these responses with observations of brain damage. Therefore, it is crucial to establish a detailed 3D organization of the brain vasculature, which could be used to develop more accurate in silico models. To this end, we have adapted our functional ultrasound imaging platform, previously designed for recording large scale activity, to enable rapid and reproducible acquisition, segmentation and reconstruction of the cortical vasculature. For the first time, it allows us to digitize the cortical \(\sim 100\)-\(\mu \)m3 spatial resolution. Unlike most available strategies, our approach can be performed in vivo within minutes. Moreover, it is easy to implement since it requires neither exogenous contrast agents nor long post-processing time. Therefore, we performed a cortex-wide reconstruction of the vasculature and its quantitative analysis, including i) classification of descending arteries versus ascending veins in more than 1500 vessels/animal and ii) rapid estimation of their length. Importantly, we confirmed the relevance of our approach in a model of cortical stroke, which allows rapid visualization of the ischemic lesion. This development contributes to extending the capabilities of ultrasound neuroimaging to better understand cerebrovascular pathologies such as stroke, vascular cognitive impairment and brain tumors, and is highly scalable for the clinic.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The availability of tools to accurately image and quantify brain vasculature is crucial for improving diagnosis, treatment, and post-treatment surveillance of various pathologies, including stroke, traumatic brain injury, vascular cognitive diseases, and brain tumors (Aziz et al., 2022). Research in ultrasound brain imaging has advanced significantly over the past decades, and several clinical applications are now emerging. In this context, \(\mu \)Doppler ultrasound imaging, which employs advanced filtering techniques compared to standard Doppler imaging, enhances sensitivity to lower velocity blood flow by reducing artifacts caused by random tissue motion. \(\mu \)Doppler imaging is based on high-frame-rate Power Doppler ultrasound (Macé et al., 2013), offering highly detailed vascular morphology. When \(\mu \)Doppler images are acquired over time using functional ultrasound (fUS) imaging, this allows for real-time detection of microvascular hemodynamics with submillimeter and subsecond precision, which correlates strongly with neuronal activity (Nunez-Elizalde et al., 2022; Lambert et al., 2024b).
\(\mu \)Doppler imaging can identify, with high resolution, key anatomical and hemodynamic features in both small animals (Brunner et al., 2020, 2021, 2023a; Bergel et al., 2018; Rabut et al., 2019), non-human primates (Norman et al., 2021; Takahashi et al., 2021b) and humans (Rabut et al., 2024; Imbault et al., 2017). Vascularity assessment is particularly important in inflammation, ischemia, injury, and tumor angiogenesis, both in peripheral organs (Tierney et al., 2019; Fu et al., 2021) and in the brain (Soloukey et al., 2023; Provost et al., 2018). A key advantage of \(\mu \)Doppler imaging is its minimally invasive nature, as it can be performed in vivo under awake conditions (Brunner et al., 2023b, 2022) and provides real-time visualization of vascular details, even at significant depths (several centimeters), without the need for contrast agents as required in CT or angio-MRI, and over a larger field-of-view (several cm) compared to two-photon imaging.
Despite the increasing availability of \(\mu \)Doppler and fUS technology in preclinical and clinical research, there are currently few tools that offer semi-automated and quantitative analysis of these images, which would be especially valuable in emergent settings where such expertise may not be readily available.
This work proposes to extract the cortex-wide vascular tree from a set of large-scale angiograms acquired using the functional ultrasound imaging modality. Existing work for complete vascular network extraction relies on dedicated angiography modalities such as magnetic resonance angiography (MRA); Hilbert et al. (2020); Tetteh et al. (2020) or optical coherence tomography (OCT); Yousefi et al. (2014); Li et al. (2017); Wu et al. (2019). The work by Cohen et al. (2018) demonstrates that vessel extraction is also feasible using ultrasound imaging, albeit for single vessels connected to a user-defined point of interest. Other work has demonstrated the extraction of the elliptical intersection of vessels from ultrasound images (Guerrero Juk et al., 2007; Wu et al., 2019). A generalized pixel profiling (Babin et al., 2012) and centerline extraction procedure (Babin et al., 2018) have been shown to robustly detect and extract vascular structures from computed tomography angiography and 3D rotational angiography images.
The ideas behind these last two works (Babin et al., 2012, 2018) are the basis of the vessel extraction in this work. The final method robustly estimates the extent to which individual voxels adhere to the properties of vessels, extracts those that are most likely to be part of vessels, and then extracts their centerline to form a 3D skeleton that allows analysis of the morphological characteristics of the blood vessel. Such processing is of interest in biomedical imaging because the morphological characteristics such as diameter, tortuosity, and shape of blood vessels are critical for early diagnosis, treatment planning, and evaluation.
Materials and Methods
The data used in this study were previously acquired and published in Brunner et al. (2023a). As the aim of the current study is to apply a novel strategy for vessel segmentation and 3D reconstruction of the cerebral vasculature based on fUS images, and not on the stroke model, only a brief explanation of the data acquisition is given. A more detailed explanation can be found in Brunner et al. (2023a).
Animals
The ultrasound imaging data used in this study originate from Brunner et al. (2023a) and are publicly available on Zenodo at the following link: https://doi.org/10.5281/zenodo.10817405. The experimental procedures were approved by the Animal Care Committee of the Catholic University of Leuven, in accordance with the national guidelines for the use of laboratory animals and the European Union Directive for animal experiments (2010/63/EU). Adult male Sprague-Dawley rats (n=9; Janvier Labs, France) with an initial weight between 200-300 g were housed in standard ventilated cages and maintained in a 12:12 hr reverse dark/light cycle environment at a temperature of 22\(^{\circ }\)C with ad libitum access to food and water. The experiments in the original study were performed only in male rats, but the aim of the study is to present the novel strategy for vessel segmentation and 3D reconstruction of the cerebral vasculature based on fUS images. Therefore, this approach to segmentation and reconstruction is not gender or age dependent.
The age and sex of the rat model may indeed influence the results on the difference of vessels detected. However,
Cranial Window for Large-scale Imaging
The surgical procedure was previously described in Brunner et al. (2023a). Briefly, the cranial window was performed in rats under isoflurane anesthesia (induction 5\(\%\), surgery 2.0-2.5\(\%\), imaging 1.5\(\%\) in compressed dry air delivered at 0.6 l/min; Iso-Vet, Dechra, Belgium). Xylocaine (0.5\(\%\), AstraZeneca, England) was injected subcutaneously into the skin of the head for preoperative analgesia. The scalp was shaved and cleaned with iso-betadine before the entire dorsal part of the skull was removed. The cranial window extended from bregma +4.0 to -7.0 mm antero-posteriorly and ± 6.0 mm from the midline. The skull was carefully removed without damaging the dura. The brain was covered with low melting 2\(\%\) agarose (Sigma-Aldrich, USA) and ultrasound gel to ensure proper acoustic coupling with the ultrasound probe.
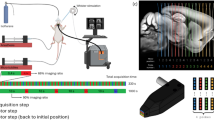
Experimental procedure for whole-brain ultrasound imaging of the rat cerebral vasculature (step 1, yellow), framework for centerline extraction and vessel skeletonization (step 2, blue), and analysis of the cortical vasculature (step 3, purple). The steps are described in detail in the Materials and Methods section
2D Scans of the Brain Vasculature with Ultrasound Imaging
The large-scale imaging procedure has been previously described in Brunner et al. (2023a). The functional ultrasound scanner is equipped with custom acquisition and processing software described by Brunner et al. (2021). The ultrasound scanner consists of a linear ultrasound transducer (15MHz, 128 elements, Xtech15, Vermon, France) connected to 128-channel transmit-receive electronics (Vantage, Verasonics, USA), both controlled by a high performance computer workstation (fUSI-2, AUTC, Estonia). The transducer was motorized (T-LSM200A, Zaber Technologies Inc., Canada) to allow antero-posterior scanning of the brain. Imaging was performed on an antivibration table to minimize external sources of vibration.
The acquisition consists of a cross-sectional coronal \(\mu \)Doppler image (12.8 mm width, 9 mm depth) composed of 300 compound images acquired at 500 Hz. Each compound image is calculated by adding nine plane waves (4.5 kHz) with angles from -12\(^{\circ }\) to 12\(^{\circ }\) with a step of 3\(^{\circ }\). The blood signal was extracted from 300 compound images using a single-value decomposition filter and removing the first 30 singular vectors (Urban et al., 2015). The \(\mu \)Doppler image is computed as the mean intensity of the blood signal in these 300 frames, which is an estimator of cerebral blood volume (CBV) Macé et al. (2011, 2013). This sequence allows a temporal resolution of 0.6s, an in-plane resolution of 100\(\times \)110\(\mu \)m, and an off-plane (image thickness) of \(~300\mu m\) Brunner et al. (2021). We performed high-resolution 2D scans of the brain vasculature consisting of 89 coronal planes from bregma (ß) +4.0 to -7.0 mm with 0.125 mm spacing (Fig. 1, step 1).
Stroke
Cortical stroke was induced by mechanical and permanent occlusion of both the left common carotid artery (CCA) and the distal branch of the left middle cerebral artery (MCA). The stroke model was established and validated by staining as described in Brunner et al. (2023a). The post-stroke 2D scan of the rat brain vasculature was performed 70 minutes after stroke onset. The ultrasound probe remained in exactly the same position between the 2 scans. All strokes were performed by the same experimenter (C.B.). The procedure to induce the cortical stroke is described in detail in Brunner et al. (2023a).
Data Processing and Analysis
The \(\mu \)Doppler image is acquired and then processed in a cascade of steps: We begin with an overview of these steps before going into more detail:
-
1)
Step 1: Acquisition of micro-Doppler images of brain vasculature (Fig. 1, Step 1).
-
a)
Optional: Color-Doppler filter to separate arteries and veins
-
a)
-
2)
Step 2: Vessel extraction and skeletonization (Fig. 1, Step 2).
-
a)
Step 2A: Detection of tubular structures using generalized R-profiling: in \(\mu \)Doppler images, vessels appear as a connected series of voxels that are brighter than their surroundings, called ridges. Generalized R-profiling finds and enhances these ridges, making them more suitable for extraction by thresholding.
-
b)
Step 2B: Filtering of segmented vessels: the enhanced ridges, which indicate vessels, are thresholded to extract foreground voxels. These foreground voxels represent the extracted vessels. As an additional filtering step, isolated foreground voxels, which are most likely to represent noise, are removed.
-
c)
Step 2C: Skeletonization of the complete brain: the centerlines of the extracted vessels are determined, and a graph type skeleton is built along these lines.
-
a)
-
3)
Step 3: Analysis of the cortical brain vasculature (Fig. 1, Step 3).
-
a)
3A: Manual specification of plane of interest: a line is drawn in the coronal cross-section \(\mu \)Doppler images of the brain indicating the location of vessels of interest. This line is extended to a plane reaching from posterior to anterior.
-
b)
3B: Extraction of vessels traversing plane of interest
-
c)
3C: Counting of the number of vessels
-
d)
3D: Analysis of vessel length distribution
-
a)
The most complex data processing steps are steps 2A and 2C, which we now explain in greater detail.
Detection of Tubular Structures using Generalized R-profiling
The goal of "Step 2A: Detection of tubular structures using generalized R-profiling" is to compute per-voxel measures that express how contrasted a voxel is compared to its multi-scale neighborhood. We achieve this by comparing the voxel’s gray value to the values found in its neighborhoods (for different sizes). This is based on the R-profiling approach described in Babin et al. (2018). The neighborhoods used in this paper are spherical structures with a radius ranging from 1 voxel to 5 voxels, chosen to correspond to the radius of the largest vessels. From each sphere of radius r, the sphere of size r-1 is subtracted to obtain a differential spherical structure. The new voxel value, called the profile measure, is the number of consecutive neighborhoods (for increasing neighborhood size) for which the current voxel is brighter than or equal to its neighborhood. For example, this brightness can be defined as the mean voxel gray value of a neighborhood. The voxel gray value of a central voxel is then compared to the mean gray value of its neighborhood of radius r=1, and if it is greater than or equal to this mean value, the process is repeated for r=2, and so on. The largest radius for which this condition is met (up to the maximum value r=5) becomes the new voxel value (Fig. 1, Step 2A).
The brightness associated with a neighborhood can be selected using a variety of basic aggregation operators on the voxel values of the neighborhood, such as mean, maximum, minimum, median, etc. The choice of these operators is made by considering both the properties of the image (such as noise and artifacts) and the nature of the structures to be extracted. It is even possible to define the operator as the average of several other operators. The selected operator is called the profile function.
Since the goal of this paper is to extract and analyze vessels, operators are defined that promote the ridge-like appearance of vessels. The different basic profile functions used are listed below:
-
1)
Maximum: the maximum value,
-
2)
Median (+): the median, taking the highest value if there are two medians,
-
3)
Median (-): the median, taking the lowest value if there are two medians,
-
4)
Minmax average: the mean of the minimum and maximum,
-
5)
Minmax root: the root of the multiplication of the minimum and maximum,
-
6)
Median root: the root of the multiplication of median (+) and median (-).
Three different operator functions were used for each rat in order to evaluate the robustness of the collection of analytical information. These three operator functions were each an average of the following three basic profile function combinations: (1,2,3), (1,4,5), and (1,5,6).
Applying these three different profile functions to a \(\mu \)Doppler image of a rat brain results in three 3D images with voxel values ranging from 0 to 5, where the detected vessel-like structures have a higher value than the non-vessel-like structures. These images are now ready to be thresholded to extract the detected vessel-like structures.
Skeleton Creation
To extract the centerline of the resulting vessels, ordered skeletonization is applied as proposed in Babin et al. (2012). For each foreground voxel, the Euclidean distance to its nearest background voxel is determined. The voxels are then sorted in ascending order by their respective distances. The voxels are iterated over in this order, and a voxel is removed if it is considered redundant to maintaining skeletal connectivity. In other words, a central voxel is redundant in its 27-neighborhood if removing it does not change the number of connected components in that neighborhood. The remaining foreground voxels represent the centerlines of the extracted vessels.
Printed version: A still image of the global procedure including rat large-scale scan, vessel extraction, and analysis. The movie can be found at https://telin.ugent.be/~astruman/cortical_reconstruction_using_fUS/Electronic version: The global procedure from large-scale scan, vessel extraction, and analysis
The extracted centerline voxels in the centerline image are subsequently labeled according to their number of foreground neighbors, yielding the following label values (Fig. 1, Step 2C):
-
0: represents an isolated voxel and is not considered further,
-
1: represents and endpoint of a vessel, and will be represented as a node in the graph-type skeleton,
-
2: represents a part of a vessel, and will be represented as a link in the graph-type skeleton,
-
>2: corresponds to a vessel bifurcation. All connected voxels with a degree larger than two represent a single vessel bifurcation. The geometric median of these voxels is used as the location of the bifurcation, thus resulting in a single node in the graph-type skeleton.
The resulting graph consists of vessel endpoints or bifurcations represented by nodes and vessel branches represented by links. Considering the three operator functions defined in the previous section, this results in three graph type skeletons. In the following sections, the quantitative results of these three operator functions are averaged out per rat.
Statistical Analysis
Comparison of vessels before and after stroke was performed by ordinary two-way ANOVA and uncorrected Fisher LSD with single pooled variance. Tests were performed with GraphPad Prism 10.0.1.
Results
In this study, we segmented the rat brain vasculature using the \(\mu \)Doppler signal acquired through functional ultrasound imaging. First, a 3D skeleton of a significant portion of the cortex in one hemisphere was automatically reconstructed Figs. 1 and 2. We then extended the analysis to identify and classify individual cortical vessels as penetrating arterioles or ascending venules based on their flow direction away from or toward the ultrasound transducer Figs. 3 and 4. Finally, we performed the 3D reconstruction of the cortical microvasculature in a pathological context of cortical stroke Fig. 5. The global procedure of large-scale scanning, vessel extraction and analysis is shown in Fig. 2.
(A) Coronal cross-section \(\mu \)Doppler image and (B) color-coded image depicting flow directionality toward (blue) or away from (red) the ultrasound probe extracted from the same brain scan. (C) Position of markers (1 to 14 from medial to lateral) used to perform the vessel extraction. (D) Front, top, lateral, and left-oblique views (top to bottom) of the 3D volume-rendered skeleton of all and (E) flow-discriminated cortical vessels. (F) Average number of cortical vessels (mean ± sem) and (G) average (top; mean ± sem, n=9) and individual distribution (bottom) of cortical vessel length. R, right; L, left; P, posterior; A, anterior; D, dorsal. Scale bar: 1mm
Ultrasound Imaging for Skeletonization of the Cortical Vasculature
Whole brain imaging was performed in head-fixed, anesthetized rats (n=9) immediately after cranial window surgery. An angiographic scan consisting of a set of 89 \(\mu \)Doppler coronal slices spaced 0.125 mm apart was acquired using a motorized ultrasound transducer (Fig. 1, step 1). From this large-scale scan, we extracted individual coronal slices of the \(\mu \)Doppler image based on the relative CBV signal (Figs. 1A and 2). We then skeletonized the brain vasculature according to the cascade of image processing steps described in the Methods section (Fig. 1, step 2). The resulting structure is a 3D skeleton of all vessels in the brain that were in the field of view of the ultrasound probe. Vessels of interest, specifically the cortical vessels of the left hemisphere, were then extracted using a user-defined plane of interest positioned below the brain surface (Figs. 1-3A, 1-3B, 3D and Fig. 2). To count the number of vessels in the extracted brain region, the plane of interest was divided into 14 sections of equal length in the medio-lateral direction. For each section, the vessels traversing its region of the plane of interest were counted (Fig. 1-3C). These vessel counts are subject to a slight error because longer vessels may pass through the plane of interest in more than one of these sections, resulting in a slight overestimation of the total number of vessels. To determine the distribution of vessel lengths, the pixel length of each vessel in the extracted skeleton of interest was calculated (Fig. 1-3D). Since the voxel size in \(\mu \)Doppler coronal images is anisotropic, this pixel length should not be interpreted as a one-to-one mapping to the actual vessel length, but rather as a correlated quantity. The exact measurement of length remains limited by the anisotropic resolution of the voxel technique used in ultrasound imaging.
Printed version: A still of the color-coded skeleton based on blood flow directionality. Arteries are indicated in red and veins in blue. The movie can be found at https://telin.ugent.be/~astruman/cortical_reconstruction_using_fUS/Electronic version: The color-coded skeleton based on blood flow directionality. Arteries are indicated in red and veins in blue
(A) Coronal cross-section \(\mu \)Doppler image extracted from the post-stroke brain scan. This coronal \(\mu \)Doppler image is at exactly the same brain location as in Fig. 2A. (B) Left-oblique view of the 3D volume-rendered skeleton of cortical vessels after stroke and average vessel loss detection along the antero-posterior axis (\(\%\) of vessel loss; color-coded line). (C) Difference in the number of cortical vessels along the medio-lateral plane of interest before (black) and after stroke (gray). Positions of the medio-lateral plane of interest showing significant differences (with a pvalue<0.001***) are highlighted in red. R, right; L, left; P, posterior; A, anterior; D, dorsal. Scale bar: 1 mm
The Doppler measurements also allow the generation of color-coded images based on blood flow direction (Figs. 3B and 4). This representation of the cerebral microvasculature allows differentiation between penetrating arteries and ascending veins in the cortex. However, the Doppler measurement is not perfectly accurate and does not provide a 3D velocity vector for the flow, but rather a projection of the vector. This still results in two separate images, one clearly showing the penetrating arteries and the other the ascending veins. These two images were processed separately in the same manner as the original \(\mu \)Doppler images (i.e., with no separation between veins and arteries). The resulting vein and artery skeletons were superimposed to visualize the combination of the two (Fig. 3E). Due to the aforementioned resolution issues, the same vessels may appear in both images, resulting in the vessel being counted as both a vein and an artery. Therefore, the total number of veins and arteries is slightly overestimated.
From this processing, we counted 892.6 ± 57.5 cortical penetrating arteries (i.e., flow direction away from the ultrasound probe; mean ± sem, n=9) and 641.7 ± 53.1 cortical ascending veins (i.e., flow direction toward the ultrasound probe; mean ± sem, n=9). We observed an uneven distribution of cortical vessels along the medio-lateral plane of interest, ranging from 114.6 ± 5.2 cortical penetrating arteries medially (i.e., position 1) to 18.0 ± 3.9 laterally (i.e., position 14; Fig. 3F, red plot) and 71.6 ± 4.4 and 23.0 ± 5.1 for ascending veins, respectively (Fig. 3F, blue plot). The medio-lateral variability can be explained by i) the uncertainty of the velocity estimator in estimating flow directionality (see zoom-in views of the color Doppler image in Fig. 3B and skeleton in (Fig. 3E), and ii) the probe-to-cortical vessel angle, which potentially affects the detection of flow directionality (Brunner et al., 2022). However, the number of penetrating and ascending vessels counted along the antero-posterior axis and between animals is very robust (Fig. 3F). In addition, the length of cortical vessels calculated from cortex-wide skeletons follows a beta distribution centered on 3-pixel-long vessels (Fig. 3G, top), representative of all rats processed (Fig. 3G, bottom).
Printed version: Still image of the skeleton of cortical vessels after stroke. The movie can be found at https://telin.ugent.be/~astruman/cortical_reconstruction_using_fUS/Electronic version: The skeleton of cortical vessels after stroke
Cortex-wide Skeletonization for Precise Detection of Ischemic Stroke
In a second and pathological application of centerline extraction, we scanned the rat brain 70 min after they were subjected to concomitant CCA+MCA occlusions and performed the skeletonization of the cortical vasculature as described above. The cortical stroke was confirmed by direct visualization of the coronal cross-section \(\mu \)Doppler images, which showed a clear loss of signal in the cortex of the left hemisphere compared to the pre-stroke scan (Fig. 3A, Supplementary Figure 1, and Figure 6). See details in Brunner et al. (2023a). In a representative case, the skeletonization of the cortical vasculature after stroke allows precise detection of the ischemic territory (Figs. 5B and 6) and shows an almost 90\(\%\) decrease in detected vessels in the center of the ischemic regions along the antero-posterior axis (Fig. 5B, color-coded line), but is significantly reduced by \(~50\%\) between lateral position 6 and 10 (gray curve, Fig. 5C and Supplementary Figure 7; pvalue<0. 001*** obtained by ordinary two-way ANOVA and uncorrected Fisher LSD with single pooled variance), i.e, where the vascular territory supplied by the distal branch of the MCA is located.
Discussion
The experimental results demonstrate that the \(\mu \)Doppler ultrasound technique generates real-time images of sufficient quality to enable the extraction of brain vasculature using the proposed skeletonization method. Our pipeline also facilitates quantitative analysis of the extracted vasculature by providing metrics such as vessel length and density, which may aid in comparing conditions between normal and pathological brains.
An important consideration in vasculature extraction is the potential overestimation of the number of vessels. This occurs due to three primary factors: i) vessels passing through multiple segments of the plane of interest, ii) spatial variations in voxel resolution inherent to ultrasound imaging techniques, and iii) uncertainty in the speed estimator, particularly when distinguishing between veins and arteries. The first factor only affects cases where the plane of interest is divided into multiple regions. This issue is avoided when the total number of vessels across the entire plane is counted, as each vessel is only counted once. One approach to mitigate overestimation in individual segments would be to count vessels only when they first traverse the plane. However, this method could reduce the accuracy of the vessel density ratio across different segments. In our experiment, we identified approximately 3000 vessels per cm\(^{3}\) in the rat cortex, consistent with previous studies utilizing two-photon imaging (Blinder et al., 2013). The potential overestimation of detected vessels could be partially mitigated by the limited spatial resolution and the exclusion of vessels with velocities below 2 mm/s, as they were filtered out using a high-pass filter to remove tissue motion. We opted to measure vessel counts in 2D rather than 3D because our primary objective was to develop a method directly applicable to 2D ultrafast Doppler images, which are more widely used compared to 3D, without the need for additional computational steps, such as multi-image fusion, to obtain complete 3D velocity profiles. Other limitations of the current approach may be addressed through improvements in hardware resolution.
At this stage, vessel lengths are given in pixels. These measurements cannot be interpreted as exact values because the pixel size is not isotropic. A solution would be to take the direction of the vessels into account when calculating their length. Knowing the exact length, width, and height of the pixels, these lengths can be converted directly into a more meaningful length metric than the number of pixels. However, in the current version, the estimation shows robustness along the brain and consistency across animals, ensuring that the results are already useful when comparing either sections of the brain or different animals.
Throughout the image processing, only one step requires user interaction: the specification of the plane of interest. In future applications, this step could be performed automatically, making the method even more robust when comparing results from different rats. For example, a brain segmentation technique could be used to find the outline of the brain, allowing the plane of interest to be automatically placed at a fixed distance from this outline. In this way, once the method has been tuned for its specific application, the process can be performed automatically without any user interaction.
In terms of resolvable vessel resolution, the data processing procedure produces results at the resolution of the functional ultrasound (fUS) image, i.e., single-voxel-wide vessels can be extracted by this procedure. The processing complexity is dominated by Step 2A. This is a numerical procedure with linear time complexity in the number of processed voxels. The pipeline we developed requires approximately 4-5 minutes per rat on a single-core CPU platform to process a 1.7-megapixel fUS image (128 px width x 150 px depth x 89 positions). Specific code optimization and implementation on a parallel computing platform (GPU / FPGA) can realistically be expected to reduce this processing time to the sub-second range, opening the door to interactive, real-time vessel imaging and quantification using the functional ultrasound imaging modality.
A direct application of centerline extraction would be to use the skeletonized large-scale vasculature as a new strategy to improve filtering, better characterize the fUS signal, and better understand cerebral hemodynamics and neurovascular coupling. In fact, one could use the skeleton as a mask to automatically extract hemodynamic parameters from veins and/or arteries, such as cerebral blood volume (CBV), red blood cell velocity, and even cerebral blood flow (CBF) in a simple and quantitative parameter. Additionally, the rapid extraction of vascular structures from uDoppler images may enhance the understanding of various vascular-related brain pathologies, such as stroke, facilitate more precise brain mapping, and inform the development of novel therapeutic strategies targeting specific vascular territories. Meanwhile, our 3D reconstruction approach combined with the recent improvement of super-resolution vascular ultrasound imaging without contrast agents (Bar-Zion et al., 2021), would allow to better separate the dense and overlapping blood vessels; thus performing a complementary analysis focusing on 2nd/3rd order arterioles, venules and capillaries, smaller than the resolution of functional ultrasound imaging modality, but where neurons mostly modulate blood supply (Rungta et al., 2021).
Moreover, the large-scale skeleton could help to improve the registration by providing a better fit with the reference atlas of the animal model (mouse, rat, monkey, ...) studied (Macé et al., 2018; Brunner et al., 2020; Takahashi et al., 2021a) either by means of vascular landmarks combined with automatic alignment approaches (Nouhoum et al., 2021) or markerless CNN-based deep learning classification (Lambert et al., 2024a).
Several strategies now allow longitudinal monitoring of brain functions in the same animal, which would be of great interest in this context when considering pathologies, as the brain vasculature and associated functions have been shown to be affected (e.g, angiogenesis, vascular rarefaction, hypertension) in several diseases including Alzheimer (Lowerison et al., 2022; Szu & Obenaus, 2021) and Parkinson diseases (Al-Bachari et al., 2020; Biju et al., 2020), tumors (Gambarota et al., 2008; Guyon et al., 2021), obesity (Pétrault et al., 2019; Gruber et al., 2021), which are currently addressed at the whole-brain level using either reduced spatiotemporal resolution technologies (Pathak et al., 2011; Lin et al., 2013) or post-mortem strategies (Todorov et al., 2020; Bumgarner & Nelson, 2022). In the context of stroke, cortex-wide skeletonization of vessels could also be used as a follow-up strategy to detect i) progressive and long-lasting neovascularization of infarcted tissue (Ergul et al., 2012; Liman & Endres, 2012), and ii) stroke-induced reverse flow (Li et al., 2010; Ergul et al., 2012), both of crucial interest when considering tissue survival and functional recovery of the insulted tissue.
Finally, this paper validates the digitization of rat brain vasculature from ultrasound images without the use of contrast agents. It presents an image processing procedure for extracting vascular structures that has been shown to be robust across different test subjects while requiring little user interaction. The entire process shows promising results with the possibility of acceleration to near real-time operation. Cortex-wide quantification in terms of vessel number and length and discrimination between veins and arteries was successfully demonstrated. The use of such quantification was demonstrated in the case of ischemic stroke, where a significant decrease in vessel number was observed in the expected region.
Information Sharing Statement
The data that was used in this study can be found on https://zenodo.org/records/10817405.
Data Availability
The data that was used in this study can be found on https://zenodo.org/records/10817405.
References
Al-Bachari, S., Naish, J., Parker, G., Emsley, H., & Parkes, L. (2020). Blood-brain barrier leakage is increased in parkinson’s disease. Frontiers in Physiology, 11
Aziz, M. U., Eisenbrey, J. R., Deganello, A., Zahid, M., Sharbidre, K., Sidhu, P., & Robbin, M. L. (2022). Microvascular flow imaging: a state-of-the-art review of clinical use and promise. Radiology, 305(2), 250–264.
Babin, D., Pizurica, A., Bellens, R., De Bock, J., Shang, Y., Goossens, B., Vansteenkiste, E., & Philips, W. (2012). Generalized pixel profiling and comparative segmentation with application to arteriovenous malformation segmentation. Medical Image Analysis, 16, 991–1002.
Babin, D., Pizurica, A., Velicki, L., Matić, V., Galić, I., Leventić, H., Zlokolica, V., & Philips, W. (2018). Skeletonization method for vessel delineation of arteriovenous malformation. Computers in Biology and Medicine, 93, 93–105.
Bar-Zion, A., Solomon, O., Rabut, C., Maresca, D., Eldar, Y., & Shapiro, M. (2021). Doppler slicing for ultrasound super-resolution without contrast agents. bioRxiv.
Bergel, A., Deffieux, T., Demené, C., Tanter, M., & Cohen, I. (2018). Local hippocampal fast gamma rhythms precede brain-wide hyperemic patterns during spontaneous rodent rem sleep. Nature communications, 9(1), 5364.
Biju, K., Shen, Q., Torres Hernandez, E., Mader, M., & Clark, R. (2020). Reduced cerebral blood flow in an \(\alpha \) -synuclein transgenic mouse model of parkinson’s disease. Journal of Cerebral Blood Flow and Metabolism, 40:0271678X1989543.
Blinder, P., Tsai, P. S., Kaufhold, J. P., Knutsen, P. M., Suhl, H., & Kleinfeld, D. (2013). The cortical angiome: an interconnected vascular network with noncolumnar patterns of blood flow. Nature Neuroscience, 16(7), 889–897.
Brunner, C., Grillet, M., Sans Dublanc, A., Farrow, K., Lambert, T., Macé, E., Montaldo, G., & Urban, A. (2020). A platform for brain-wide volumetric functional ultrasound imaging and analysis of circuit dynamics in awake mice. Neuron, 108
Brunner, C., Grillet, M., Urban, A., Roska, B., Montaldo, G., & Macé, E. (2021). Whole-brain functional ultrasound imaging in awake head-fixed mice. Nature Protocols, 16
Brunner, C., Lagumersindez Denis, N., Gertz, K., Grillet, M., Montaldo, G., Endres, M., & Urban, A. (2023a). Brain-wide continuous functional ultrasound imaging for real-time monitoring of hemodynamics during ischemic stroke. Journal of Cerebral Blood Flow and Metabolism, 0(0), 0271678X231191600. PMID: 37503862.
Brunner, C., Macé, E., Montaldo, G., & Urban, A. (2022). Quantitative hemodynamic measurements in cortical vessels using functional ultrasound imaging. Frontiers in Neuroscience, 16, 831650.
Brunner, C., Montaldo, G., & Urban, A. (2023b). Functional ultrasound imaging of stroke in awake rats. eLife, 12:RP88919.
Bumgarner, J., & Nelson, R. (2022). Open-source analysis and visualization of segmented vasculature datasets with vesselvio. Cell reports methods.
Cohen, E., Deffieux, T., Demené, C., Cohen, L., & Tanter, M. (2018). 3d vessel extraction in the rat brain from ultrasensitive doppler images. Lecture Notes in Bioengineering, pages 81–91
Ergul, A., Alhusban, A., & Fagan, S. C. (2012). Angiogenesis. Stroke, 43(8), 2270–2274.
Fu, Z., Zhang, J., Lu, Y., Wang, S., Mo, X., He, Y., Wang, C., & Chen, H. (2021). Clinical applications of superb microvascular imaging in the superficial tissues and organs: a systematic review. Academic Radiology, 28(5), 694–703.
Gambarota, G., Leenders, W., Maass, C., Wesseling, P., Van der Kogel, A., Tellingen, O., & Heerschap, A. (2008). Characterisation of tumor vasculature in mouse brain by uspio contrast-enhanced mri. British Journal of Cancer, 98, 1784–9.
Gruber, T., Pan, C., Contreras, R., Wiedemann, T., Morgan, D., Skowronski, A., Lefort, S., Murat, C., Le Thuc, O., Legutko, B., Ruiz-Ojeda, F., Fuente-Fernández, M., García-Villalón, A., González-Hedström, D., Huber, M., szigeti buck, K., Müller, T., Ussar, S., Pfluger, P., & García Cáceres, C. (2021). Obesity-associated hyperleptinemia alters the gliovascular interface of the hypothalamus to promote hypertension. Cell Metabolism, 33
Guerrero Juk, J., Salcudean, S., Mcewen, J., Masri, B., & Nicolaou, S. (2007). Real-time vessel segmentation and tracking for ultrasound imaging applications. IEEE Transactions on Medical Imaging, 26, 1079–90.
Guyon, J., Chapouly, C., Andrique, L., Bikfalvi, A., & Daubon, T. (2021). The normal and brain tumor vasculature: Morphological and functional characteristics and therapeutic targeting. Frontiers in Physiology, page 622615
Hilbert, A., Madai, V., Akay, E., Aydin, O., Behland, J., Sobesky, J., Galinovic, I., Khalil, A., Taha, A. A., Wuerfel, J., Dusek, P., Niendorf, T., Fiebach, J., Frey, D., & Livne, M. (2020). Brave-net: Fully automated arterial brain vessel segmentation in patients with cerebrovascular disease. Frontiers in artificial intelligence.
Imbault, M., Chauvet, D., Gennisson, J.-L., Capelle, L., & Tanter, M. (2017). Intraoperative functional ultrasound imaging of human brain activity. Scientific Reports, 7(1), 7304.
Lambert, T., Brunner, C., Kil, D., Wuyts, R., D’Hondt, E., Montaldo, G., & Urban, A. (2024). A deep learning classification task for brain navigation in rodents using micro-doppler ultrasound imaging. Heliyon, 10(5), e27432.
Lambert, T., Niknejad, H. R., Kil, D., Brunner, C., Nuttin, B., Montaldo, G., & Urban, A. (2024b). Functional ultrasound imaging and neuronal activity: how accurate is the spatiotemporal match? bioRxiv, pages 2024–07
Li, A., You, J., Du, C., & Pan, Y. (2017). Automated segmentation and quantification of oct angiography for tracking angiogenesis progression. Biomedical Optics Express, 8, 5604.
Li, L., Ke, Z., Tong, K. Y., & Ying, M. (2010). Evaluation of cerebral blood flow changes in focal cerebral ischemia rats by using transcranial doppler ultrasonography. Ultrasound in Medicine and Biology, 36(4), 595–603.
Liman, T., & Endres, M. (2012). New vessels after stroke: Postischemic neovascularization and regeneration. Cerebrovascular diseases (Basel, Switzerland), 33, 492–9.
Lin, C.-Y., Siow, T., Lin, M.-H., Hsu, Y.-H., Tung, Y.-Y., Jang, T., Recht, L., & Chang, C. (2013). Visualization of rodent brain tumor angiogenesis and effects of antiangiogenic treatment using 3d r2-mra. Angiogenesis, 16
Lowerison, M., Chandra Sekaran, N., Zhang, W., Dong, Z., Chen, X., Llano, D., & Song, P. (2022). Aging-related cerebral microvascular changes visualized using ultrasound localization microscopy in the living mouse. Scientific Reports, 12.
Macé, E., Montaldo, G., Cohen, I., Baulac, M., Fink, M., & Tanter, M. (2011). Functional ultrasound imaging of the brain. Nature Methods, 8, 662–4.
Macé, E., Montaldo, G., Osmanski, B.-F., Cohen, I., Fink, M., & Tanter, M. (2013). Functional ultrasound imaging of the brain: Theory and basic principles. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 60, 92–506.
Macé, E., Montaldo, G., Trenholm, S., Cowan, C., Brignall, A., Urban, A., & Roska, B. (2018). Whole-brain functional ultrasound imaging reveals brain modules for visuomotor integration. Neuron, 100, 1241-1251.e7.
Norman, S. L., Maresca, D., Christopoulos, V. N., Griggs, W. S., Demene, C., Tanter, M., Shapiro, M. G., & Andersen, R. A. (2021). Single-trial decoding of movement intentions using functional ultrasound neuroimaging. Neuron, 109(9), 1554–1566.
Nouhoum, M., Ferrier, J., Osmanski, B.-F., Ialy-Radio, N., Pezet, S., Tanter, M., & Deffieux, T. (2021). A functional ultrasound brain gps for automatic vascular-based neuronavigation. Scientific Reports, 11, 15197.
Nunez-Elizalde, A. O., Krumin, M., Reddy, C. B., Montaldo, G., Urban, A., Harris, K. D., & Carandini, M. (2022). Neural correlates of blood flow measured by ultrasound. Neuron, 110(10), 1631–1640.
Pathak, A., Kim, E., Zhang, J., & Jones, M. (2011). Three-dimensional imaging of the mouse neurovasculature with magnetic resonance microscopy. PloS one, 6, e22643.
Provost, J., Garofalakis, A., Sourdon, J., Bouda, D., Berthon, B., Viel, T., Perez-Liva, M., Lussey-Lepoutre, C., Favier, J., Correia, M., amp, et al. (2018). Simultaneous positron emission tomography and ultrafast ultrasound for hybrid molecular, anatomical and functional imaging. Nature Biomedical Engineering, 2(2), 85–94.
Pétrault, O., Petrault, M., Ouk, T., Bordet, R., Berezowski, V., & Bastide, M. (2019). Visceral adiposity links cerebrovascular dysfunction to cognitive impairment in middle-aged mice. Neurobiology of Disease, 130, 104536.
Rabut, C., Correia, M., Finel, V., Pezet, S., Pernot, M., Deffieux, T., & Tanter, M. (2019). 4d functional ultrasound imaging of whole-brain activity in rodents. Nature methods, 16(10), 994–997.
Rabut, C., Norman, S. L., Griggs, W. S., Russin, J. J., Jann, K., Christopoulos, V., Liu, C., Andersen, R. A., & Shapiro, M. G. (2024). Functional ultrasound imaging of human brain activity through an acoustically transparent cranial window. Science Translational Medicine, 16(749), eadj3143
Rungta, R., Zuend, M., Aydin, A.-K., Martineau, E., Boido, D., Weber, B., & Charpak, S. (2021). Diversity of neurovascular coupling dynamics along vascular arbors in layer ii/iii somatosensory cortex. Communications Biology, 4, 855.
Soloukey, S., Verhoef, L., Generowicz, B. S., De Zeeuw, C. I., Koekkoek, S. K., Vincent, A. J., Dirven, C. M., Harhangi, B. S., & Kruizinga, P. (2023). Case report: High-resolution, intra-operative \(\mu \)doppler-imaging of spinal cord hemangioblastoma. Frontiers in Surgery, 10, 1153605.
Szu, J., & Obenaus, A. (2021). Cerebrovascular phenotypes in mouse models of alzheimer’s disease. Journal of Cerebral Blood Flow and Metabolism : Official Journal of the International Society of Cerebral Blood Flow and Metabolism, 41, 271678X21992462
Takahashi, D., Hady, A., Zhang, Y., Liao, D., Montaldo, G., Urban, A., & Ghazanfar, A. (2021a). Social-vocal brain networks in a non-human primate. bioRxiv.
Takahashi, D. Y., El Hady, A., Zhang, Y. S., Liao, D. A., Montaldo, G., Urban, A., & Ghazanfar, A. A. (2021b). Social-vocal brain networks in a non-human primate. BioRxiv, pages 2021–12.
Tetteh, G., Efremov, V., Forkert, N. D., Schneider, M., Kirschke, J., Weber, B., Zimmer, C., Piraud, M., & Menze, B. (2020). Deepvesselnet: Vessel segmentation, centerline prediction, and bifurcation detection in 3-d angiographic volumes. Frontiers in Neuroscience, 14.
Tierney, J., Walsh, K., Griffith, H., Baker, J., Brown, D. B., & Byram, B. (2019). Combining slow flow techniques with adaptive demodulation for improved perfusion ultrasound imaging without contrast. IEEE Transactions on Ultrasonics, Ferroelectrics, and Frequency Control, 66(5), 834–848.
Todorov, M., Paetzold, J., Schoppe, O., Tetteh, G., Shit, S., Efremov, V., Todorov-Völgyi, K., Düring, M., Dichgans, M., Piraud, M., Menze, B., & Ertürk, A. (2020). Machine learning analysis of whole mouse brain vasculature. Nature Methods, 17, 1–8.
Urban, A., Brunner, C., Dussaux, C., Chassoux, F., Devaux, B., & MONTALDO, G. (2015). Functional ultrasound imaging of cerebral capillaries in rodents and humans. Jacobs Journal of Molecular and Translational Medicine, 1, 007.
Wu, C., Xie, Y., Shao, L., Yang, J., Ai, D., Song, H., Wang, Y., & Huang, Y. (2019). Automatic boundary segmentation of vascular doppler optical coherence tomography images based on cascaded u-net architecture. OSA Continuum, 2, 677.
Yousefi, S., Liu, T., & Wang, R. (2014). Segmentation and quantification of blood vessels for oct-based micro-angiograms using hybrid shape/intensity compounding. Microvascular Research, 97
Acknowledgements
We thank all NERF animal caretakers, including I. Eyckmans, F. Ooms, and S. Luijten, for their help with the management of the animals.
Funding
This work is supported by grants from KU Leuven (C14/18/099-STYMULATE-STROKE). The functional ultrasound imaging platform is supported by grants from FWO (G091719N, 1197818N, G0A4F24N, G055124N), ERANET, EU Horizon 2020 (Grant number 964215, UnscrAMBLY), HORIZON-MSCA-2022-DN-01 (Project 101119916 - SOPRANI). Théo Lambert is funded by IMEC PhD Talent grant, Clément Brunner is funded by a FWO Senior Postdoctoral fellowship (12D7523N). Anoek Strumane was partly funded by the Research Foundation - Flanders, through its Strategic Basic Research Programme (FWO-SBO) project PASCELL (parameter-activated sorting of cells - S001423N).
Author information
Authors and Affiliations
Contributions
Conceptualization was done by T.L., D.B., C.B. and A.U. The methodology was designed by A.S., T.L., J.A., D.B., C.B. and A.U. The software was developed by A.S., D.B., G.M. and A.U. Animal preparation and planning was done by C.B. Data analysis was performed by A.S., J.A. and D.B. Visualizations were created by A.S. and C.B. All authors contributed to the writing and reviewing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
Ethics approval was received by the KU Leuven ethical committee with the ethics approval number ECD P095/2017.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Appendix
Appendix
Related to Figure 3A-B. \(\mu \)Doppler scans of the cortical vasculature before and after stroke. Scale bar: 1mm
Related to Figure 3C. Number of cortical vessels along the medio-lateral plane of interest before (black) and after stroke (grey) for every single rat used in this work
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Strumane, A., Lambert, T., Aelterman, J. et al. Large Scale in vivo Acquisition, Segmentation and 3D Reconstruction of Cortical Vasculature using \(\mu \)Doppler Ultrasound Imaging. Neuroinform 23, 5 (2025). https://doi.org/10.1007/s12021-024-09706-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s12021-024-09706-1