Abstract
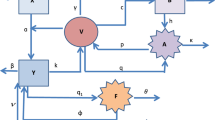
Dengue disease is caused by an infected mosquito bite and manifests in different clinical symptoms. The complexity of the pathogenesis of dengue virus and the limitations of biological knowledge have been barriers to completely understanding the progress of this disease. To address this concern, we developed a mathematical model of the immune response to eliminate dengue virus. The model considered both cellular and humoral immune responses, and we evaluated their contributions to the clearance of dengue virus.We also performed global sensitivity analysis and parameters estimation using clinical data. We found the global stability for virus-free equilibrium and for the virus-presence equilibrium, we concluded that to avoid oscillations in the model and to control the viral load, a strong proliferation of cytotoxic cells must prevail. However, if there exists a weak proliferation of cytotoxic cells, the way to avoid instabilities is to either inhibit the differentiation of T-CD4+ helper cells in Th1 cells or increase the proliferation of B cells.






Similar content being viewed by others
References
Alberts, B., et al.: Molecular Biology of the Cell, 4th edn. Garland Science, New York (2002)
Ansari, H., Hesaaraki, M.: A with-in host dengue infection model with immune response and beddington-deangelis incidence rate. Appl. Math. 3(02), 177 (2012)
Ben-Shachar, R., Koelle, K.: Minimal within-host dengue models highlight the specific roles of the immune response in primary and secondary dengue infections. J. R. Soc. Interface 12(103), 20140886 (2015)
Cerón Gómez, M., Yang, H.M.: A simple mathematical model to describe antibody-dependent enhancement in heterologous secondary infection in dengue. Math. Med. Biol. J. IMA 36, 411–438 (2019)
Chen, Y.-C., Wang, S.-Y.: Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J. Virol. 76(19), 9877–9887 (2002)
Clapham, H.E., et al.: Within-host viral dynamics of dengue serotype 1 infection. J. R. Soc. Interface 11(96), 20140094 (2014)
Courageot, M.-P., Catteau, A., Despres, P.: Mechanisms of dengue virus-induced cell death. Adv. Virus Res. 60, 157–186 (2003)
de Matos, A.M., et al.: CD8+ T lymphocyte expansion, proliferation and activation in dengue fever. PLoS Negl. Trop. Diseases 9(2), e0003520–e0003520 (2015)
Esteva, L., Yang, H.M.: Assessing the effects of temperature and dengue virus load on dengue transmission. J. Biol. Syst. 23(4), 527–554 (2015)
Gantmacher, F.R., Brenner, J.L.: Applications of the Theory of Matrices. Dover Publications, New York (2005)
Gujarati, T.P., Ambika, G.: Virus antibody dynamics in primary and secondary dengue infections. J. Math. Biol. 69(6–7), 1773–1800 (2014)
Hellerstein, M., et al.: Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat. Med. 5(1), 83–89 (1999)
Hwang, T.-W., Wang, F.-B.: Dynamics of a dengue fever transmission model with crowding effect in human population and spatial variation. Discrete Contin. Dyn. Syst. Ser. B 18(1), 147–161 (2013)
Karl, S., Halder, N., Kelso, J.K., Ritchie, S.A., Milne, G.J.: A spatial simulation model for dengue virus infection in urban areas. BMC Infectious Dis. 14(1), 447 (2014)
Knipe, D.M., Howley, P.M., Griffin, D., Lamb, R., Martin, M., Roizman, B., Straus, S.: Fields Virology. Lippincott Williams & Wilkins, Philadelphia (2001)
Kou, Z., et al.: Human antibodies against dengue enhance dengue viral infectivity without suppressing type i interferon secretion in primary human monocytes. Virology 410(1), 240–247 (2011)
Macdonald, G.: The analysis of equilibrium in malaria. Trop. Dis. Bull. 49(9), 813–829 (1952)
Mak, T.W., Saunders, M.E.: The Immune Response: Basic and Clinical Principles. Academic Press, Berlin (2005)
Mandell, G., Bennett, J.E., Dolin, R.: Principles and Practices of Infectious Diseases, 7th edn. Churchill Livingstone Elsevier, Philadelphia (2010)
Marianneau, P., Flamand, M., Deubel, V., Desprès, P.: Induction of programmed cell death (apoptosis) by dengue virus in vitro and in vivo. Acta Cient. Venez. 49, 13–17 (1997)
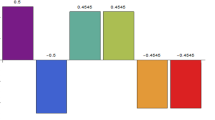
Marino, S., et al.: A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 254(1), 178–196 (2008)
Michalewicz, Z.: Genetic Algorithms + Data Structures= Evolution Programs, 3rd edn. Springer, Berlin (1996)
Myint, K.S., et al.: Cellular immune activation in children with acute dengue virus infections is modulated by apoptosis. J. Infect. Dis. 194(5), 600–607 (2006)
Nikin-Beers, R., Ciupe, S.M.: The role of antibody in enhancing dengue virus infection. Math. Biosci. 263, 83–92 (2015)
Nuraini, N., et al.: A with-in host dengue infection model with immune response. Math. Comput. Model. 49(5), 1148–1155 (2009)
Palmer, D.R., et al.: Differential effects of dengue virus on infected and bystander dendritic cells. J. Virol. 79(4), 2432–2439 (2005)
Perera, S., Perera, S.: Simulation model for dynamics of dengue with innate and humoral immune responses. In: Computational and Mathematical Methods in Medicine (2018)
Ribeiro, R.M., et al.: In vivo dynamics of T cell activation, proliferation, and death in HIV-1 infection: Why are CD4+ but not CD8+ T cells depleted? Proc. Natl. Acad. Sci. 99(24), 15572–15577 (2002)
Sasmal, S.K., Dong, Y., Takeuchi, Y.: Mathematical modeling on t-cell mediated adaptive immunity in primary dengue infections. J. Theor. Biol. 429, 229–240 (2017)
Sithisarn, P., et al.: Behavior of the dengue virus in solution. J. Med. Virol. 71(4), 532–539 (2003)
Sun, P., et al.: CD40 ligand enhances dengue viral infection of dendritic cells: a possible mechanism for T cell-mediated immunopathology. J. Immunol. 177(9), 6497–6503 (2006)
Yang, H.M.: The basic reproduction number obtained from jacobian and next generation matrices-a case study of dengue transmission modelling. Biosystems 126, 52–75 (2014)
Yang, H.M.: A mathematical model to assess the immune response against trypanosoma cruzi infection. J. Biol. Syst. 23(01), 131–163 (2015)
Yang, H.M.: The transovarial transmission in the dynamics of dengue infection: epidemiological implications and thresholds. Math. Biosci. 286, 1–15 (2017)
Yang, H.M., Boldrini, J.L., Fassoni, A.C., Freitas, L.F.S., Gomez, M.C., de Lima, K.K.B., Andrade, V.R., Freitas, A.R.R.: Fitting the incidence data from the city of Campinas, Brazil, based on dengue transmission modellings considering time-dependent entomological parameters. PLoS ONE 11(3), e0152186 (2016)
Yang, H.M., Macoris, M.L.G., Galvani, K.C., Andrighetti, M.T.M., Wanderley, D.M.V.: Assessing the effects of temperature on dengue transmission. Epidemiol. Infect. 137(08), 1179–1187 (2009)
Acknowledgements
This study was supported by research Grant 2013/17264-0, São Paulo Research Foundation (FAPESP)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendices
1.1 A Genetic Algorithm
The function to be minimized is \(f(V,\varUpsilon )=(V-V_{data})^{2}\), where V is the viral load, given by positive solution of the system (1)–( 4) at steady state, \(V_{data}\) are the data of viral load of patients and \( \varUpsilon =(\beta _{_{I}},\alpha _{v},\gamma _{1},\gamma _{2},\alpha _{r},\alpha _{a},\alpha _{cr},\alpha _{ca},\mu _{v})\) is the set of the unknown parameters, where \(\beta _{_{I}}\in (\beta _{_{I}min},\beta _{_{I}max})\), \(\alpha _{v}\in (\alpha _{vmin},\alpha _{vmax})\), \(\ldots \), \( \mu _{v}\in (\mu _{vmin},\mu _{vmax})\). The first step is the transformation of each parameter in binary and form a string called chromosome. Let \( \varGamma \) be the binary representation of \(\varUpsilon \). Then
will be the chromosome, which has just 1’s and 0’s and \(\beta _{_{I}2}\), \(\alpha _{v2}\), \(\gamma _{12}\), \(\gamma _{22}\), \(\alpha _{r2}\), \(\alpha _{a2} \), \(\alpha _{cr2}\), \(\alpha _{ca2}\ \text {and}\ \mu _{v2}\) are the binary representation of parameters. This chromosome has length \( m=\sum _{i=1}^{9}m_{i}\), where \(m_{1}\) is the smallest integer such that \( (\beta _{_{I}max}-\beta _{_{I}min})\times 10^{p}<2^{m_{1}}-1\), \(m_{2}\) is the smallest integer such that \((\alpha _{vmax}-\alpha _{vmin})\times 10^{p}<2^{m_{2}}-1\), \(\dots \), \(m_{9}\) is the smallest integer such that \( (\mu _{vmax}-\mu _{vmin})\times 10^{p}<2^{m_{9}}-1\) and p is the number indicating decimal places desirable for the parameters. Each \(m_{i}\) is the length of binary string of parameters.
The algorithm is:
-
1.
Initial population
We create a random population \(P_{0}\) of chromosomes, where each chromosome is a binary string of length \(m=\sum _{i=1}^{9}m_{i}\). We suppose that this initial population has n chromosomes, i.e.,
$$\begin{aligned} P_{0}=\{\varGamma _{0}^{1},\ldots ,\varGamma _{0}^{n}\}. \end{aligned}$$ -
2.
Evaluation of function
At this step we evaluate the function f at each element of the population \( P_{0}\), that is, \(f(V,\varUpsilon ^{i})\), where \(\varUpsilon ^{i}\) is the decimal representation of \(\varGamma _{0}^{i}\), \(i=1\), \(\dots \), n.
-
3.
Next Population
At this step we select the next population by applying the genetic operator (crossover and mutations).
-
Selection method
In order to select the population, we apply the tournament selection method, which consists in selecting randomly some number k of chromosome and storing the minimum of the set \(\{f(V,\varUpsilon ^{J_{1}})\), \(\ldots \), \(f(V, \varUpsilon ^{J_{k}})\}\) of k elements, where J is a subset of k elements (\(J\subset \{1,2,\ldots ,n\}\)), into the next generation. This process is repeated n times. Obviously, some chromosomes would be selected more than once. Now, we apply the crossover and mutations operators to this selected population.
-
Crossover operator
This operator apply recombination in the chromosomes (see Fig. 7). We give the probability of crossover \(p_{c}\). This probability gives us the expected number \(p_{c}\times n\) of chromosomes, which undergo the crossover operation. The process of crossover function is done in the following way: for each chromosome in the (new) population, we generate a random number r from the range [0, 1]. If \(r<p_{c}\), we select this chromosome for crossover.
If the number of selected chromosomes is even, we can pair them easily. If the number of selected chromosomes were odd, we would either add one extra chromosome or remove one selected chromosome, which is made randomly as well. The operator explained here is known as one-point crossover. There are other crossover operator as: two-point crossover, uniform crossover and half uniform crossover.
-
Mutation operator
This operator applies alterations in the elements of chromosomes (changes of 0 for 1 and vice versa (see Fig. 8). We give the probability of mutation \(p_{m}\). This probability gives us the expected number of mutated elements \( p_{m}\times m\times _{n}\). The process to perform the mutation operator is similar to the crossover operator: for each chromosome in the current (i.e., after crossover) population and for each element within the chromosome, a random number r is generated in the range [0, 1]. If \(r<p_{m}\), then we mutate the element.
-
-
4.
After all the above steps, we have created the first generation: population \(P_{1}\). Now just repeat the steps 2 and 3 to \(P_{1}\), and the process goes up to the desired generations.
A detailed explanation of genetic algorithms can be found in [22]. The algorithm adapted and used in the simulations can be accessed in the link http://people.csail.mit.edu/gbezerra/Code/GA/ga.m.
Rights and permissions
About this article
Cite this article
Gómez, M.C., Yang, H.M. Mathematical model of the immune response to dengue virus. J. Appl. Math. Comput. 63, 455–478 (2020). https://doi.org/10.1007/s12190-020-01325-8
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12190-020-01325-8
Keywords
- Dengue virus
- Adaptive immune response
- Humoral and cellular immune responses
- Deterministic model
- Net reproduction number of virus