Abstract
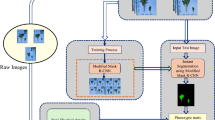
In agriculture, plant phenotyping serves as a critical process for assessing a wide range of plant traits that are pivotal for crop management and improvement, such as plant height, leaf area, flowering time, and resistance to diseases. Traditional methods employed in plant phenotyping often encounter significant challenges, including time-consuming procedures, inefficiencies in data processing, and vulnerabilities to environmental variability. These limitations hinder the accuracy and scalability required for effective agricultural practices. To address these issues, this paper introduces the Hybrid U Mask Regional Convolutional Pelican Search (HUMRC-PS) method as an innovative approach in plant phenotyping. This method leverages advanced technologies to overcome the shortcomings of conventional techniques. Specifically, HUMRC-PS utilizes mask Region-based Convolutional Neural Networks (RCNN), a sophisticated deep learning architecture, to accurately segment plant areas within images. By precisely delineating plant features from complex backgrounds, RCNN enables a focused analysis of key attributes such as leaf morphology, color variations, and overall size. Furthermore, the integration of U-net, another deep learning framework, enhances the method’s capacity to capture both local and global features from plant images. This capability is crucial for comprehensive trait measurement and analysis, as it ensures that nuanced details and broader characteristics are accounted for in the phenotyping process. Moreover, HUMRC-PS incorporates Pelican optimization with a crossover strategy to fine-tune its internal parameters and optimize model performance. This approach not only enhances the accuracy and efficiency of plant trait identification but also contributes to the method’s adaptability across different agricultural scenarios and environmental conditions. Through rigorous experimentation on an image dataset tailored for plant phenotyping, the study validates the effectiveness of HUMRC-PS. It evaluates the method’s performance using a spectrum of evaluation metrics, demonstrating superior results with 98.76% accuracy when compared to existing methodologies. This validation underscores HUMRC-PS’s potential to significantly advance plant phenotyping practices by providing more precise, efficient, and scalable solutions. By surpassing the limitations of traditional approaches, HUMRC-PS offers promising opportunities for improving agricultural productivity, disease management, and breeding programs through enhanced understanding and characterization of plant traits.















Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Afrakhteh S, Mosavi MR, Khishe M, Ayatollahi A (2020) Accurate classification of EEG signals using neural networks trained by hybrid population-physic-based algorithm. Int J Autom Comput 17(1):108–122
Arya S, Sandhu KS, Singh J, Kumar S (2022) Deep learning: as the new frontier in high-throughput plant phenotyping. Euphytica 218(4):47
Azimi S, Kaur T, Gandhi TK (2021) A deep learning approach to measure stress level in plants due to nitrogen deficiency. Measurement 173:108650
Cardellicchio A, Solimani F, Dimauro G, Petrozza A, Summerer S, Cellini F, Renò V (2023) Detection of tomato plant phenotyping traits using YOLOv5-based single stage detectors. Comput Electron Agric 207:107757
Das Choudhury S, Guha S, Das A, Das AK, Samal A, Awada T (2022) Flowerphenonet: automated flower detection from multi-view image sequences using deep neural networks for temporal plant phenotyping analysis. Remote Sens 14(24):6252
ElManawy AI, Sun D, Abdalla A, Zhu Y, Cen H (2022) HSI-PP: a flexible open-source software for hyperspectral imaging-based plant phenotyping. Comput Electron Agric 200:107248
Ghosh S, Singh A, Kumar S (2023) BBBC-U-Net: optimizing U-Net for automated plant phenotyping using big bang big crunch global optimization algorithm. Int J Inf Technol 15:4375–4387
Gill T, Gill SK, Saini DK, Chopra Y, de Koff JP, Sandhu KS (2022) A comprehensive review of high throughput phenotyping and machine learning for plant stress phenotyping. Phenomics 2(3):156–183
Gupta S, Deep K (2019) Improved sine cosine algorithm with crossover scheme for global optimization. Knowl Based Syst 165:374–406
Hati AJ, Singh RR (2021) Artificial intelligence in smart farms: plant phenotyping for species recognition and health condition identification using deep learning. AI 2(2):274–289
Hati AJ, Singh RR (2023) AI-driven pheno-parenting: a deep learning based plant phenotyping trait analysis model on a novel soilless farming dataset. IEEE Access 11:35298–35314
Kalsotra R, Arora S (2023) Performance analysis of U-Net with hybrid loss for foreground detection. Multimed Syst 29(2):771–786
Kaveh M, Khishe M, Mosavi MR (2019) Design and implementation of a neighborhood search biogeography-based optimization trainer for classifying sonar dataset using multi-layer perceptron neural network. Analog Integr Circuits Signal Process 100:405–428
Khishe M, Mohammadi H (2019) Passive sonar target classification using multi-layer perceptron trained by salp swarm algorithm. Ocean Eng 181:98–108
Khishe M, Mosavi MR (2019) Improved whale trainer for sonar datasets classification using neural network. Appl Acoust 154:176–192
Khishe M, Mosavi MR (2020) Classification of underwater acoustical dataset using neural network trained by chimp optimization algorithm. Appl Acoust 157:107005
Khishe M, Safari A (2019) Classification of sonar targets using an MLP neural network trained by dragonfly algorithm. Wirel Pers Commun 108(4):2241–2260
Khishe M, Mosavi MR, Kaveh M (2017) Improved migration models of biogeography-based optimization for sonar dataset classification by using neural network. Appl Acoust 118:15–29
Khishe M, Mosavi MR, Moridi A (2018) Chaotic fractal walk trainer for sonar data set classification using multi-layer perceptron neural network and its hardware implementation. Appl Acoust 137:121–139
Koh JC, Spangenberg G, Kant S (2021) Automated machine learning for high-throughput image-based plant phenotyping. Remote Sens 13(5):858
Li Y, Yang Z (2017) Application of EOS-ELM with binary Jaya-based feature selection to real-time transient stability assessment using PMU data. IEEE Access 5:23092–23101
Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenotyping. Sensors 14(11):20078–20111
Li D, Shi G, Kong W, Wang S, Chen Y (2020) A leaf segmentation and phenotypic feature extraction framework for multiview stereo plant point clouds. IEEE J Sel Top Appl Earth Obs Remote Sens 13:2321–2336
Li J, Zhang D, Yang F, Zhang Q, Pan S, Zhao X, Zhang Q, Han Y, Yang J, Wang K, Zhao C (2024) TrG2P: a transfer learning-based tool integrating multi-trait data for accurate prediction of crop yield. Plant Commun 5(7):100975
Liu H, Bruning B, Garnett T, Berger B (2020) Hyperspectral imaging and 3D technologies for plant phenotyping: from satellite to close-range sensing. Comput Electron Agric 175:105621
Ma X, Zhu K, Guan H, Feng J, Yu S, Liu G (2019) High-throughput phenotyping analysis of potted soybean plants using colorized depth images based on a proximal platform. Remote Sens 11(9):1085
Mahmoodzadeh A, Nejati HR, Mohammadi M, Ibrahim HH, Khishe M, Rashidi S, Ali HFH (2022) Prediction of Mode-I rock fracture toughness using support vector regression with metaheuristic optimization algorithms. Eng Fract Mech 264:108334
Mosavi MR, Khishe M (2017) Training a feed-forward neural network using particle swarm optimizer with autonomous groups for sonar target classification. J Circuits Syst Comput 26(11):1750185
Mosavi MR, Khishe M, Akbarisani M (2017) Neural network trained by biogeography-based optimizer with chaos for sonar data set classification. Wirel Pers Commun 95:4623–4642
Mosavi MR, Khishe M, Naseri MJ, Parvizi GR, Mehdi AYAT (2019) Multi-layer perceptron neural network utilizing adaptive best-mass gravitational search algorithm to classify sonar dataset. Arch Acoust 44(1):137–151
Mousavipour F, Mosavi MR (2023) Sonar data classification using neural network trained by hybrid dragonfly and chimp optimization algorithms. Wirel Pers Commun 129(1):191–208
Nabiee S, Harding M, Hersh J, Bagherzadeh N (2022) Hybrid U-Net: semantic segmentation of high-resolution satellite images to detect war destruction. Mach Learn Appl 9:100381
Okyere FG, Cudjoe D, Sadeghi-Tehran P, Virlet N, Riche AB, Castle M, Greche L, Mohareb F, Simms D, Mhada M, Hawkesford MJ (2023) Machine learning methods for automatic segmentation of images of field-and glasshouse-based plants for high-throughput phenotyping. Plants 12(10):2035
Pineda M, Barón M, Pérez-Bueno ML (2020) Thermal imaging for plant stress detection and phenotyping. Remote Sens 13(1):68
Pound MP, Atkinson JA, Townsend AJ, Wilson MH, Griffiths M, Jackson AS, Bulat A, Tzimiropoulos G, Wells DM, Murchie EH, Pridmore TP (2017) Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 6(10):gix083
Qiao W, Khishe M, Ravakhah S (2021) Underwater targets classification using local wavelet acoustic pattern and multi-layer perceptron neural network optimized by modified whale optimization algorithm. Ocean Eng 219:108415
Saffari A, Khishe M, Zahiri SH (2022) Fuzzy-ChOA: an improved chimp optimization algorithm for marine mammal classification using artificial neural network. Analog Integr Circuits Signal Process 111(3):403–417
Salehi B, Mireei SA, Jafari M, Hemmat A, Majidi MM (2024) Integrating in-field Vis-NIR leaf spectroscopy and deep learning feature extraction for growth-stage dependent and independent genotyping of wheat plants. Biosyst Eng 238:188–199
Tariq M, Ahmed M, Iqbal P, Fatima Z, Ahmad SL (2020) Crop phenotyping. Systems modeling. Springer, Singaprore, pp 45–60
Trojovský P, Dehghani M (2022) Pelican optimization algorithm: a novel nature-inspired algorithm for engineering applications. Sensors 22(3):855
Wang H, Duan Y, Shi Y, Kato Y, Ninomiya S, Guo W (2021) EasyIDP: a Python package for intermediate data processing in UAV-based plant phenotyping. Remote Sens 13(13):2622
Wang K, Abid MA, Rasheed A, Crossa J, Hearne S, Li H (2023) DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants. Mol Plant 16(1):279–293
Xiao Q, Bai X, Zhang C, He Y (2022) Advanced high-throughput plant phenotyping techniques for genome-wide association studies: a review. J Adv Res 35:215–230
Zhang Y, Li T, Na G, Li G (2015) Li Y (2015) Optimized extreme learning machine for power system transient stability prediction using synchrophasors. Math Probl Eng 1:529724
Zhang Q, Chang X, Bian SB (2020) Vehicle-damage-detection segmentation algorithm based on improved mask RCNN. IEEE Access 8:6997–7004
Zhang C, Yi Y, Wang L, Zhang X, Chen S, Su Z, Zhang S, Xue Y (2024) Estimation of the bio-parameters of winter wheat by combining feature selection with machine learning using multi-temporal unmanned aerial vehicle multispectral images. Remote Sens 16(3):469
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Human and animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, P., Senthilselvi, A., Manju, I. et al. HUMRC-PS: Revolutionizing plant phenotyping through Regional Convolutional Neural Networks and Pelican Search Optimization. Evolving Systems 15, 2211–2230 (2024). https://doi.org/10.1007/s12530-024-09612-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12530-024-09612-6