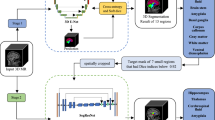
Abstract
Segmenting the whole brain into a large number (for example, \(\ge 100\)) of regions is challenging due to the complexity of the brain and the lack of annotated data. Deep neural network-based segmentation methods have shown promise, but due to the limitation of graphics processing unit (GPU) memory, they cannot fully exploit the brain structure information contained in 3D data. This paper proposes a memory-efficient framework to exploit the global brain structure for whole-brain segmentation. In this framework, upon extracting the brain region by using a skull-stripping subnetwork, a global modeling subnetwork is used to learn a global brain representation for segmentation, while an adaptable segmentation subnetwork is used to optimize the global representation during training and directly segment the whole brain during testing. This framework enables the representation to be learned from the global structure with reduced memory consumption, and segmentation is performed without splitting the brain into patches. To overcome the lack of annotated data, we also propose a semi-supervised method based on a symmetry consistency loss and a prior knowledge-based pseudolabel generation strategy. Extensive experiments on four datasets demonstrate that our method outperforms previously developed methods and achieves state-of-the-art performance. The method is computationally efficient in that segmenting a raw magnetic resonance imaging (MRI) image requires less than 2 s on a TITAN X GPU; our approach is much faster than multiatlas-based methods and previously proposed 3D deep learning methods. The code is publicly available at https://github.com/ZYX-MLer/AGNetwork.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Landman BA, Warfield SK. MICCAI 2012: Workshop on multi-atlas labeling. In: MICCAI.
Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-atlas segmentation with joint label fusion. TPAMI. 2013;35(3):611–23.
Wang H, Yushkevich P. Multi-atlas segmentation with joint label fusion and corrective learning-an open source implementation. Front Neuroinform. 2013;7:27.
Sun L, Shao W, Wang M, Zhang D, Liu M. High-order feature learning for multi-atlas based label fusion: Application to brain segmentation with MRI. TMI. 2020;29:2702–13.
Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. 2015;108:214–24.
Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJNL, Išgum I. Automatic segmentation of MR brain images with a convolutional neural network. TMI. 2016;35(5):1252–61.
Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in mri images. TMI. 2016;35(5):1240–51.
Hussain S, Anwar SM, Majid M. Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing. 2018;282:248–61.
de Brébisson A, Montana G. Deep neural networks for anatomical brain segmentation. In: CVPRW. 2015. p. 20–28.
Mehta R, Majumdar A, Sivaswamy J. Brainsegnet: a convolutional neural network architecture for automated segmentation of human brain structures. J Med Imaging. 2017;4(2).
Guha Roy A, Conjeti S, Navab N, Wachinger C. Quicknat: A fully convolutional network for quick and accurate segmentation of neuroanatomy. NeuroImage. 2019;186:713–727.
Ganaye P-A, Sdika M, Triggs B, Benoit-Cattin H. Removing segmentation inconsistencies with semi-supervised non-adjacency constraint. MIA. 2019;58:101551.
Li Y, Li H, Fan Y. Acenet: Anatomical context-encoding network for neuroanatomy segmentation. MIA. 2021;70:101991.
Roy AG, Navab N, Wachinger C. Recalibrating fully convolutional networks with spatial and channel “squeeze and excitation” blocks. TMI. 2019;38(2):540–9.
Roy AG, Conjeti S, Sheet D, Katouzian A, Navab N, Wachinger C. Error corrective boosting for learning fully convolutional networks with limited data. In: MICCAI. Springer; 2017. p. 231–239.
Dai C, Mo Y, Angelini E, Guo Y, Bai W. Transfer learning from partial annotations for whole brain segmentation. In: DART, MIL3ID. Springer; 2019. p. 199–206.
Wachinger C, Reuter M, Klein T. Deepnat: Deep convolutional neural network for segmenting neuroanatomy. NeuroImage. 2018;170:434–445. Segmenting the Brain.
Huo Y, Xu Z, Xiong Y, Aboud K, Parvathaneni P, Bao S, Bermudez C, Resnick SM, Cutting LE, Landman BA. 3d whole brain segmentation using spatially localized atlas network tiles. NeuroImage. 2019;194:105–19.
Coupé P, Mansencal B, Clément M, Giraud R, Denis de Senneville B, Ta V-T, Lepetit V, Manjon JV. Assemblynet: A large ensemble of CNNS for 3D whole brain MRI segmentation. NeuroImage. 2020;219:117026.
Bontempi D, Benini S, Signoroni A, Svanera M, Muckli L. Cerebrum: a fast and fully-volumetric convolutional encoder-decoder for weakly-supervised segmentation of brain structures from out-of-the-scanner MRI. MIA. 2020;62:101688.
Zhao Y-X, Zhang Y-M, Song M, Liu C-L. Multi-view semi-supervised 3D whole brain segmentation with a self-ensemble network. In MICCAI. Springer; 2019. p. 256–265.
Zhang Y, Liu B, Wang Y, Gao Z, Bai X, Zhou F. Bdb-Net: Boundary-enhanced dual branch network for whole brain segmentation. In MICCAI; 2020. pp. 188–197, Springer.
Sun L, Ma W, Ding X, Huang Y, Liang D, Paisley J. A 3D spatially weighted network for segmentation of brain tissue from MRI. TMI. 2020;39(4):898–909.
Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: MICCAI. Springer; 2016. p. 424–432.
Paschali M, Gasperini S, Roy AG, Fang MY-S, Navab N. 3DQ: compact quantized neural networks for volumetric whole brain segmentation. In: MICCAI. Springer; 2019. p. 438–446.
Li W, Wang G, Fidon L, Ourselin S, Cardoso MJ, Vercauteren T. On the compactness, efficiency, and representation of 3D convolutional networks: Brain parcellation as a pretext task. In: Information Processing in Medical Imaging (M. Niethammer, M. Styner, S. Aylward, H. Zhu, I. Oguz, P.-T. Yap, and D. Shen, eds.), (Cham); 2017. pp. 348–360, Springer International Publishing.
Li Y, Cui J, Sheng Y, Liang X, Wang J, Chang EI-C, Xu Y. Whole brain segmentation with full volume neural network. Comput Med Imaging Graph. 2021;93:101991.
Ouali Y, Hudelot C, Tami M. Semi-supervised semantic segmentation with cross-consistency training. In CVPR; 2020. pp. 12671–12681.
Chen X, Yuan Y, Zeng G, Wang J. Semi-supervised semantic segmentation with cross pseudo supervision. In CVPR; 2021. pp. 2613–2622.
Lai X, Tian Z, Jiang L, Liu S, Zhao H, Wang L, Jia J. Semi-supervised semantic segmentation with directional context-aware consistency. In CVPR; 2021. pp. 1205–1214.
Huang R, Noble JA, Namburete AI. Omni-supervised learning: scaling up to large unlabelled medical datasets. In MICCAI; 2018. pp. 572–580, Springer.
Cui W, Liu Y, Li Y, Guo M, Li Y, Li X, Wang T, Zeng X, Ye C. Semi-supervised brain lesion segmentation with an adapted mean teacher model. In IPMI; 2019. pp. 554–565, Springer.
Nie D, Gao Y, Wang L, Shen D. Asdnet: Attention based semi-supervised deep networks for medical image segmentation. In MICCAI; 2018. pp. 370–378, Springer.
Zhang Y, Yang L, Chen J, Fredericksen M, Hughes DP, Chen DZ. Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In MICCAI; 2017. pp. 408–416, Springer.
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In MICCAI; 2015. pp. 234–241, Springer.
Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. TPAMI. 2017;39(4):640–51.
Ulyanov D, Vedaldi A, Lempitsky V. Instance normalization: The missing ingredient for fast stylization, arXiv preprint arXiv:1607.08022, 2016.
He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In ICCV; 2015. pp. 1026–1034.
Van Engelen JE, Hoos HH. A survey on semi-supervised learning. Mach Learn. 2020;109(2):373–440.
Lin T-Y, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. TPAMI. 2020;42(2):318–27.
Milletari F, Navab N, Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 3DV; 2016. pp. 565–571.
Zhou Z, Sodha V, Siddiquee MMR, Feng R, Tajbakhsh N, Gotway MB, and J. Liang, Models genesis: Generic autodidactic models for 3d medical image analysis. In MICCAI; 2019. pp. 384–393, Springer.
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, et al. Pytorch: An imperative style, high-performance deep learning library. NIPS. 2019;32:8026–37.
Klein A, Dal Canton T, Ghosh SS, Landman B, Lee J, Worth A. Open labels: online feedback for a public resource of manually labeled brain images. In OHBM; 2010. vol. 84358.
Kennedy DN, Haselgrove C, Hodge SM, Rane PS, Makris N, Frazier JA. Candishare: a resource for pediatric neuroimaging data. Neuroinformatics. 2012; pp. 319–322.
Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19(9):1498–507.
Jack CR, Bernstein MA, Fox NC, Thompson P, Weiner MW. The alzheimer’s disease neuroimaging initiative (adni): Mri methods. J Magn Reson Imaging; 2010. vol. 27, no. 4, pp. 685–691.
Tarvainen A, Valpola H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In NIPS; 2017. vol. 30, p. 1195–1204, Inc.
Sajjadi M, Javanmardi M, Tasdizen T. Regularization with stochastic transformations and perturbations for deep semi-supervised learning. NIPS. 2016;29:1163–71.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. In NIPS; 2017. pp. 5998–6008.
Funding
This work has been supported by the National Key Research and Development Program Grant 2018AAA0100400, the National Natural Science Foundation of China (NSFC) grants 61773376, 61836014, 61721004, and 31870984.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, YX., Zhang, YM., Song, M. et al. Adaptable Global Network for Whole-Brain Segmentation with Symmetry Consistency Loss. Cogn Comput 14, 2246–2259 (2022). https://doi.org/10.1007/s12559-022-10011-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-022-10011-9