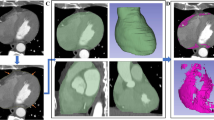
Abstract
Epicardial adipose tissue (EAT) is contiguous with arteries and myocardium. An increase in the volume of EAT may lead to adverse cardiovascular events. Therefore, quantification of EAT is necessary. The purpose of this paper is to employ a more than helpful algorithm for EAT segmentation and quantification. First, we used a simple convolutional neural network to select EAT slices, which significantly reduced oversegmentation. Then, we employed multiscale residual attention Unet (MRA-Unet) to achieve EAT segmentation based on the selected slices. Finally, we calculated the segmented volume to quantify EAT. We used 33/103 patients to test the model. The average Dice score for EAT segmentation was 0.883. For EAT quantification, the Pearson and concordance correlation coefficients reached 0.973 and 0.971, respectively. The results showed that our algorithm had strong agreement and consistency with expert. Our method performed efficient quantification and had strong consistency and agreement with the volume manually marked by experts. This algorithm can be used as a tool to assist in the clinical quantification of EAT. By combining different measurements to predict adverse cardiovascular and heart disease events, it has the potential to be applied for clinical use in the future.









Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Madonna R, Massaro M, Scoditti E, Pescetelli I, De Caterina R. The epicardial adipose tissue and the coronary arteries: dangerous liaisons. Cardiovasc Res. 2019;115:1013–25.
Greulich S, Maxhera B, Vandenplas G, de Wiza DH, Smiris K, Mueller H, et al. Secretory products from epicardial adipose tissue of patients with type 2 diabetes mellitus induce cardiomyocyte dysfunction. Circulation. 2012;126:2324–34.
Franssens BT, Nathoe HM, Leiner T, van der Graaf Y, Visseren FL. Relation between cardiovascular disease risk factors and epicardial adipose tissue density on cardiac computed tomography in patients at high risk of cardiovascular events. Eur J Prev Cardiol. 2017;24:660–70.
Tanaka K, Fukuda D, Sata M. Roles of epicardial adipose tissue in the pathogenesis of coronary atherosclerosis - an update on recent findings. Circ J. 2020;85:2–8.
Packer M. Epicardial adipose tissue may mediate deleterious effects of obesity and inflammation on the myocardium. J Am Coll Cardiol. 2018;71:2360–72.
Nattel S. Aguilar M. Electrophysiological effects of atrial epicardial adipose tissue: keep your friends close and your enemies closer. J Am Coll Cardiol. 2020;1212–1214.
Zhao L, Harrop DL, Ng ACT, Wang WYS. Epicardial adipose tissue is associated with left atrial dysfunction in people without obstructive coronary artery disease or atrial fibrillation. Can J Cardiol. 2018;34:1019–25.
Parisi V, Rengo G, Perrone-Filardi P, Pagano G, Femminella GD, Paolillo S, et al. Increased epicardial adipose tissue volume correlates with cardiac sympathetic denervation in patients with heart failure. Circ Res. 2016;118:1244–53.
Kitagawa T, Nakamoto Y, Fujii Y, Sasaki K, Tatsugami F, Awai K, et al. Relationship between coronary arterial (18)F-sodium fluoride uptake and epicardial adipose tissue analyzed using computed tomography. Eur J Nucl Med Mol Imaging. 2020;47:1746–56.
White IA. Cardiac sympathetic denervation in the failing heart: a role for epicardial adipose tissue. Circ Res. 2016;1189–1191.
Shan D, Wang X, Dou G, Zhang W, Jing J, He B, et al. Vascular-specific epicardial adipose tissue in predicting functional myocardial ischemia for patients with stable chest pain. J Thromb Thrombolysis. 2021;51:915–23.
Payne GA, Kohr MC, Tune JD. Epicardial perivascular adipose tissue as a therapeutic target in obesity-related coronary artery disease. Br J Pharmacol. 2012;165:659–69.
Mahabadi AA, Lehmann N, Kälsch H, Robens T, Bauer M, Dykun I, et al. Association of epicardial adipose tissue with progression of coronary artery calcification is more pronounced in the early phase of atherosclerosis: results from the Heinz Nixdorf recall study. JACC Cardiovasc Imaging. 2014;7:909–16.
de Vos AM, Prokop M, Roos CJ, Meijs MFL, van der Schouw YT, Rutten A, et al. Peri-coronary epicardial adipose tissue is related to cardiovascular risk factors and coronary artery calcification in post-menopausal women. Eur Heart J. 2008;29:777–83.
Yerramasu A, Dey D, Venuraju S, Anand DV, Atwal S, Corder R, et al. Increased volume of epicardial fat is an independent risk factor for accelerated progression of sub-clinical coronary atherosclerosis. Atherosclerosis. 2012;220:223–30.
Le Jemtel TH, Samson R, Ayinapudi K, Singh T, Oparil S. Epicardial adipose tissue and cardiovascular disease. Curr Hypertens Rep. Current Hypertension Reports 2019;21.
Militello C, Rundo L, Toia P, Conti V, Russo G, Filorizzo C et al. A semi-automatic approach for epicardial adipose tissue segmentation and quantification on cardiac CT scans. Comput Biol Med [Internet]. Elsevier Ltd; 2019;114:103424. Available from: https://doi.org/10.1016/j.compbiomed.2019.103424.
Zlokolica V, Krstanović L, Velicki L, Popović B, Janev M, Obradović R, et al. Semiautomatic epicardial fat segmentation based on fuzzy c-means clustering and geometric ellipse fitting. J Healthc Eng. 2017;2017:5817970.
Dey D, Suzuki Y, Suzuki S, Ohba M, Slomka PJ, Polk D, et al. Automated quantitation of pericardiac fat from noncontrast CT. Invest Radiol. 2008;43:145–53.
de Albuquerque VHC, de A Rodrigues D, Ivo RF, Peixoto SA, Han T, Wu W et al. Fast fully automatic heart fat segmentation in computed tomography datasets. Comput Med Imaging Graph. 2020;80:101674.
Ding X, Terzopoulos D, Diaz-Zamudio M, Berman DS, Slomka PJ, Dey D. Automated epicardial fat volume quantification from non-contrast CT. In: Ourselin S, Styner MA, editors. Med Imaging Image Process. 2014;90340I.
Shahzad R, Bos D, Metz C, Rossi A, Kirisli H, van der Lugt A, et al. Automatic quantification of epicardial fat volume on non-enhanced cardiac CT scans using a multi-atlas segmentation approach. Med Phys. 2013;40:91910.
Ismael AM, Şengür A. The investigation of multiresolution approaches for chest X-ray image based COVID-19 detection. Heal Inf Sci Syst. Springer International Publishing. 2020;8:1–11.
Commandeur F, Goeller M, Betancur J, Cadet S, Doris M, Chen X, et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans Med Imaging. 2018;37:1835–46.
He X, Guo BJ, Lei Y, Wang T, Fu Y, Curran WJ, et al. Automatic segmentation and quantification of epicardial adipose tissue from coronary computed tomography angiography. Phys Med Biol. 2020;65:95012.
Zhang Q, Zhou J, Zhang B, Member S, Jia W. Automatic epicardial fat segmentation and quantification of CT scans using dual U-Nets with a morphological processing layer. IEEE Access. 2020:1–10.
Commandeur F, Goeller M, Razipour A, Cadet S, Hell MM, Kwiecinski J, et al. Fully automated CT quantification of epicardial adipose tissue by deep learning: a multicenter study. Radiol Artif Intell. 2019;1: e190045.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Med Image Comput Comput Interv – MICCAI 2015. Cham: Springer International Publishing; 2015. p. 234–41.
Huang H, Lin L, Tong R, Hu H, Zhang Q, Iwamoto Y et al. UNet 3+: a full-scale connected UNet for medical image segmentation. ICASSP 2020 - 2020 IEEE Int Conf Acoust Speech Signal Process. 2020;1055–1059.
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J, et al. UNet++: a nested U-Net architecture for medical image segmentation. In: Stoyanov D, Taylor Z, Carneiro G, Syeda-Mahmood T, Martel A, Maier-Hein L, et al., editors. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support. Cham: Springer International Publishing; 2018. p. 3–11.
Tamarappoo B, Dey D, Shmilovich H, Nakazato R, Gransar H, Cheng VY, et al. Increased pericardial fat volume measured from noncontrast CT predicts myocardial ischemia by SPECT. JACC Cardiovasc Imaging. 2010;3:1104–12.
Rahman T, Akinbi A, Chowdhury MEH, Rashid TA, Şengür A, Khandakar A et al. COV-ECGNET: COVID-19 detection using ECG trace images with deep convolutional neural network. Heal Inf Sci Syst. Springer International Publishing. 2022;10:1–16.
Dou Q, Chen H, Yu L, Qin J, Heng P-A. Multilevel contextual 3-D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans Biomed Eng. 2017;64:1558–67.
Ami A, Ae B, Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst Appl. 2020;164.
Ramachandran P, Zoph B, Le QV. Swish: a self-gated activation function. arXiv Neural Evol Comput. 2017.
Gomes JC, Barbosa VA d. F, Santana MA, Bandeira J, Valença MJS, de Souza RE et al. IKONOS: an intelligent tool to support diagnosis of COVID-19 by texture analysis of X-ray images. Res Biomed Eng. 2020.
Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. In: Gordon G, Dunson D, Dudík M, editors. Proc Fourteenth Int Conf Artif Intell Stat [Internet]. Fort Lauderdale, FL, USA: PMLR 2011;315–23. Available from: http://proceedings.mlr.press/v15/glorot11a.html.
Pratiwi H, Perdana Windarto A, Susliansyah S, Restu Aria R, Susilowati S, Kanti Rahayu L et al. Sigmoid activation function in selecting the best model of artificial neural networks. J Phys Conf Ser. 2020;12010.
Fang Z, Chen Y, Nie D, Lin W, Shen D. RCA-U-Net: residual channel attention U-Net for fast tissue quantification in magnetic resonance fingerprinting. Med Image Comput Comput Assist Interv. 2019;11766:101–9.
Zhang Y, Li K, Li K, Wang L, Zhong B, Fu Y. Image super-resolution using very deep residual channel attention networks. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Comput Vis – ECCV 2018. Cham: Springer International Publishing; 2018. p. 294–310.
Wang P, Chen P, Yuan Y, Liu D, Huang Z, Hou X et al. Understanding convolution for semantic segmentation. IEEE Winter Conf Appl Comput Vis. 2018;1451–60.
Chen L-C, Papandreou G, Schroff F, Adam H. Rethinking atrous convolution for semantic image segmentation. arXiv e-prints. 2017 arXiv:1706.05587.
Kingma DP, Ba JL. Adam: a method for stochastic optimization. 3rd Int Conf Learn Represent ICLR Conf Track Proc. 2015;1–15.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. IEEE Conf Comput Vis Pattern Recognit. 2017;2261–2269.
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. IEEE Conf Comput Vis Pattern Recognit. 2016;2818–2826.
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T et al. MobileNets: efficient convolutional neural networks for mobile vision applications 2017. arXiv e-prints. arXiv:1704.04861.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2016;770–778.
Hu J, Shen L, Sun G. Squeeze-and-excitation networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit IEEE. 2018;7132–7141.
Milletari F, Navab N, Ahmadi S-A. V-Net: fully convolutional neural networks for volumetric medical image segmentation Fourth Int Conf 3D Vis. 2016;565–571.
Qin X, Zhang Z, Huang C, Dehghan M, Zaiane OR, Jagersand M. U2-Net: going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020;106.
Yang T, Zhou Y, Li L, Zhu C. DCU-Net: Multi-scale U-Net for brain tumor segmentation. J Xray Sci Technol. 2020;28:709–26.
Tan M, Le Q. EfficientNet: rethinking model scaling for convolutional neural networks. In: Chaudhuri K, Salakhutdinov R, editors. Proc 36th Int Conf Mach Learn [Internet]. PMLR 2019;6105–6114. Available from: https://proceedings.mlr.press/v97/tan19a.html.
Zhao Y, Han R, Rao Y. A new feature pyramid network for object detection. Proc Int Conf Virtual Real Intell Syst ICVRIS. 2019;428–431.
Ni Z-L, Bian G-B, Zhou X-H, Hou Z-G, Xie X-L, Wang C, et al. RAUNet: residual attention U-Net for semantic segmentation of cataract surgical instruments. In: Gedeon T, Wong KW, Lee M, editors., et al., Neural Inf Process. Cham: Springer International Publishing; 2019. p. 139–49.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 81871380, 81771909, and 62171300).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical Approval
A non-contrast CT dataset, from 103 patients, has been collected at the Beijing Chaoyang Hospital of Capital Medical University, and the ethics committee of Beijing Chaoyang Hospital approved this study.
Conflict of Interest
The authors declare no competing interests.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qu, J., Chang, Y., Sun, L. et al. Deep Learning-Based Approach for the Automatic Quantification of Epicardial Adipose Tissue from Non-Contrast CT. Cogn Comput 14, 1392–1404 (2022). https://doi.org/10.1007/s12559-022-10036-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-022-10036-0