Abstract
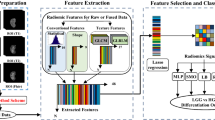
Grading of glioma is crucial for both treatment decisions and prognosis assessments. This study proposes a fast, simple, and accurate prediction framework for the non-invasive grading of glioma based on radiomics. The framework consists of four main steps. First, glioma images were subjected to semi-automatic segmentation to reduce the heavy workload. Then, 346 radiomics features were calculated from the segmented regions of interest. However, selecting features directly from such a large set to train the prediction model might lead to overfitting. Therefore, a de-redundancy algorithm was proposed to construct a candidate feature set based on mutual information. Finally, feature selection was executed using elastic net, and a grading model with linear regression was built. The proposed non-invasive solution for the grading of glioma can potentially hasten treatment decision, with the use of a de-redundancy algorithm that significantly improved the prediction accuracy. Experiments were conducted on 161 glioma samples from Henan Provincial People’s Hospital between 2012 and 2016, and results demonstrated the accurate grading effect and the generality of the de-redundancy algorithm. Moreover, the proposed framework exhibited desirable sensitivity (93.57%), specificity (86.53%), AUC (0.9638) and accuracy (91.30%).







Similar content being viewed by others
Notes
ITK is an open-source, cross-platform system that provides developers with an extensive suite of software tools for image analysis. More details in https://itk.org/.
References
Aerts HJ et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006. https://doi.org/10.1038/ncomms5006
Amadasun M, King R (1989) Textural features corresponding to textural properties. Syst Man Cybern IEEE Trans 19:1264–1274
Bishop CM (1995) Neural networks for pattern recognition. Agric Eng Int CIGR J Sci Res Dev Manuscr Pm 12:1235–1242
Bocchino C, Carabellese A, Caruso T, Della Sala G, Ricart S, Spinella A (1990) Use of gray value distribution of run lengths for texture analysis. Pattern Recogn Lett 11:415–419
Bradley PS, Mangasarian OL (1998) Feature selection via concave minimization and support vector machines. In: Fifteenth international conference on machine learning, pp 82–90
Cai D, Zhang C, He X (2010) Unsupervised feature selection for multi-cluster data. In: ACM SIGKDD international conference on knowledge discovery and data mining, pp 333–342
Chaddad A, Zinn PO (2015) Colen RR Radiomics texture feature extraction for characterizing GBM phenotypes using GLCM. In: IEEE international symposium on biomedical imaging
Cover TM, Thomas JA (1991) Elements of information theory. J Am Stat Assoc 39:1600–1601
Dasarathy BV, Holder EB (1991) Image characterizations based on joint gray level—run length distributions. Pattern Recogn Lett 12:497–502
Dean BL, Drayer BP, Bird CR, Flom RA, Hodak JA, Coons SW, Carey RG (1990) Gliomas: classification with MR imaging. Radiology 174:411–415
Echegaray S, Gevaert O, Shah R, Kamaya A, Louie J, Kothary N, Napel S (2015) Core samples for radiomics features that are insensitive to tumor segmentation: method and pilot study using CT images of hepatocellular carcinoma. J Med Imaging. https://doi.org/10.1117/1.Jmi.2.4.041011
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1
Galloway MM (1975) Texture analysis using gray level run lengths. Comput Graph Image Process 4:172–179
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Gu Y et al (2013) Automated delineation of lung tumors from CT images using a single click ensemble segmentation approach. Pattern Recogn 46:692–702
Guyon I (2001) Erratum: gene selection for cancer classification using support vector machines. Mach Learn 46:389–422
Hall MA (1999) Correlation-based feature selection for machine learning. PhD Thesis, Waikato Univer-sity, 19
Haralick RM, Shanmugam K, Dinstein IH (1973) Textural features for image classification. IEEE Trans Syst Man Cybern SMC3:610–621
He X, Cai D, Niyogi P (2006) Laplacian score for feature selection. In: International conference on neural information processing systems, pp 507–514
Hsieh KL, Lo CM, Hsiao CJ (2017) Computer-aided grading of gliomas based on local and global MRI features. Comput Methods Progr Biomed 139:31
Huang Y-Q (2015) Development and validation of a radiomics nomogram for preoperative predicting lymph node metastasis in colorectal cancer. J Clin Oncol. https://doi.org/10.1200/JCO.2015.65.9128
Huang YQ et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157. https://doi.org/10.1200/Jco.2015.65.9128
Hunter RD (1980) WHO handbook for reporting results of cancer treatment British. J Cancer 38:484–485
Hutter M (2002) Robust feature selection using distributions of mutual information
Li Q, Griffiths JG (2004) Least squares ellipsoid specific fitting. In: Geometric modeling and processing, proceedings, pp 335–335
Li-Chun Hsieh K, Chen CY, Lo CM (2017) Quantitative glioma grading using transformed gray-scale invariant textures of MRI. Comput Biol Med 83:102–108. https://doi.org/10.1016/j.compbiomed.2017.02.012
Liu H, Motoda H (2007) Computational methods of feature selection. CRC Press, Boca Raton
Lloyd SP (1982) Least squares quantization in PCM. IEEE Trans Inf Theory 28:129–137
Ma J, Wang Q, Ren Y, Hu H, Zhao J (2016) Automatic lung nodule classification with radiomics approach. In: Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations, vol 9789, pp 978906. https://doi.org/10.1117/12.2220768
Macyszyn L et al (2016) Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-oncology 18:417–425. https://doi.org/10.1093/neuonc/nov127
Nie K et al (2016) Rectal cancer: assessment of neoadjuvant chemo-radiation outcome based on radiomics of multi-parametric MRI. Clin Cancer Res Off J Am Assoc Cancer Res
Nyul LG, Udupa JK, Zhang X (2000) New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 19:143–150
Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS (2016) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro-Oncology 18:v1–v75. https://doi.org/10.1093/neuonc/now207
Oubel E, Beaumont H, Iannessi A (2016) Mutual information-based feature selection for radiomics. 9789:97890L. https://doi.org/10.1117/12.2216746
Peng H, Ding C, Long F (2015) Minimum redundancy—maximum relevance feature selection feature selection
Qin JB et al (2017) Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences medical science monitor international medical. J Exp Clin Res 23:2168–2178
Roffo G, Melzi S (2016) Feature selection via eigenvector centrality. In: New frontiers in mining complex patterns in conjunction with Ecml/pkdd
Roffo G, Melzi S, Cristani M (2015) Infinite feature selection. In: International conference on computer vision, pp 4202–4210
Roffo G, Melzi S, Castellani U, Vinciarelli A (2017) Infinite latent feature selection: a probabilistic latent graph-based ranking approach. In: IEEE international conference on computer vision
Skogen K, Schulz A, Dormagen JB, Ganeshan B, Helseth E, Server A (2016) Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 85:824–829. https://doi.org/10.1016/j.ejrad.2016.01.013
Smith SM et al (2004) Advances in functional and structural MR image analysis and implementation as. FSL Neuroimage 23:208–219
Therasse P et al (2000) New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst 87:881–886
Thibault G et al. (2009) Texture indexes and gray level size zone matrix application to cell nuclei classification pattern recognition and information processing (PRIP)
Vallières M, Freeman CR, Skamene SR, El Naqa I (2015) A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 60:5471–5496
van Klaveren RJ, Aerts JGJV., de Bruin H, Giaccone G, Manegold C, van Meerbeeck JP (2004) Inadequacy of the RECIST criteria for response evaluation in patients with malignant pleural mesothelioma. Lung Cancer 43:63–69. https://doi.org/10.1016/s0169-5002(03)00292-7
Veeramuthu A, Meenakshi S, Darsini VP (2015) Brain image classification using learning machine approach and brain structure analysis. Proc Comput Sci 50:388–394. https://doi.org/10.1016/j.procs.2015.04.030
Weston J, Elisseeff A, Schlkopf B, Tipping M (2003) Use of the zero-norm with linear models and kernel methods. J Mach Learn Res 3:1439–1461
Wu WM et al. (2016) Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. https://doi.org/10.3389/Fonc.2016.00071
Wu YP, Lin YS, Wu WG, Yang C, Gu JQ, Bai Y, Wang MY (2017) Semiautomatic segmentation of glioma on mobile devices. J Healthc Eng 2017:8054939. https://doi.org/10.1155/2017/8054939
Yang Y, Shen HT, Ma Z, Huang Z, Zhou X (2011) l 2,1-norm regularized discriminative feature selection for unsupervised learning. In: International joint conference on artificial intelligence, pp 1589–1594
Yoon HJ et al. (2015) Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine. https://doi.org/10.1097/Md.0000000000001753
Yu J et al (2016) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. https://doi.org/10.1007/s00330-016-4653-3
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency reliability. Neuroimage 31:1116–1128
Zeng H, Cheung YM (2011) Feature selection and kernel learning for local learning-based clustering. IEEE Trans Pattern Anal Mach Intell 33:1532–1547
Acknowledgements
This study was funded by National Natural Science Foundation of China (Grants 81772009, 91630206, 91330117, and 81720108021), the National Key Research and Development Program of China (Grant 2016YFB0201800), the China Postdoctoral Science Foundation (Grant 2016M590948), Social development, science and technology research projects in Shaanxi Province (Grant 2016SF-428), Henan Province Scientific and Technological Innovation Talents Project (Grant 164200510014), Henan Province Scientific and Technological Cooperation Project (Grant 152106000014).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, Y., Liu, B., Wu, W. et al. Grading glioma by radiomics with feature selection based on mutual information. J Ambient Intell Human Comput 9, 1671–1682 (2018). https://doi.org/10.1007/s12652-018-0883-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-018-0883-3