Abstract
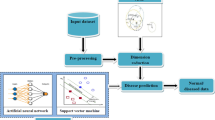
A good dimensional reduction technique is needed to apply and improve the effectiveness of dimensionality reduction for medical data. High-dimensional data brings great challenges in terms of computational complexity and classification efficiency. It is necessary to compress effectively from high dimensional space to low dimensional space to design a learning curve with good performance. Therefore, dimensional reduction is necessary to study and understand the mechanism of the practical problems of medical data. In this paper, a hybrid local fisher discriminant analysis (HLFDA) method is proposed for the dimension reduction of the medical data. LFDA is a localized variant of Fisher discriminant analysis and it is popular for supervised dimensionality reduction method. The proposed HLFDA is a combination of Locality-preserving projection and LFDA. After the dimensionality reduction process, the data are given to the Type2fuzzy neural network classifier to classify a given data as normal or abnormal. The paper focused on improving performance in terms of prediction accuracy. Three types of UCI cancer dataset is used for analyzing the performance of the proposed method.


















Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.References
Alshamlan H, Badr G, Alohali Y (2015) mRMR-ABC: a hybrid gene selection algorithm for cancer classification using microarray gene expression profiling. Biomed Res Int 2015:1–15
Anagaw A, Chang YL (2019) A new complement naïve Bayesian approach for biomedical data classification. J Ambient Intell Human Comput 10(10):3889–3897
Aziz R, Verma CK, Srivastava N (2018) Artificial neural network classification of high dimensional data with novel optimization approach of dimension reduction. Ann Data Sci 5(4):615–635
Cascianelli S, Bello-Cerezo R, Bianconi F, Fravolini ML, Belal M, Palumbo B, Kather JN (2018) Dimensionality reduction strategies for cnn-based classification of histopathological images. In: 2018 Int Conf on intell interact multimed syst and serv, pp 21–30
Cheriguene S, Azizi N, Dey N, Ashour AS, Mnerie CA, Olariu T, Shi F(2016) Classifier ensemble selection based on mRMR algorithm and diversity measures: an application of medical data classification. In: 2016 Int workshop soft comput appl, pp 375–384
Cho JH, Kim DH (2011) intelligent feature selection by bacterial foraging algorithm and information theory. In: 2011 Int conf on adv commun and netw, pp 238–244
Fu L (2015) The discriminate analysis and dimension reduction methods of high dimension. Open J Soc Sci 3(03):7–13
Ghalwash MF, Ramljak D, Obradović Z (2015) Patient-specific early classification of multivariate observations. Int J Data Mining Bioinform 11(4):392–411
Gupta K, Janghel RR (2019) Dimensionality reduction-based breast cancer classification using machine learning. In: Computational intelligence: theories, applications and future directions, pp 133–146
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Indra P, Manikandan M (2020) Multilevel Tetrolet transform based breast cancer classifier and diagnosis system for healthcare applications. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-01755-z
Kinal M, Woźniak M (2020) Data preprocessing for des-knn and its application to imbalanced medical data classification. In: 2020 Asian conf on intell inf and database syst, pp 589–599
Krishnakumar S, Manivannan K (2020) Effective segmentation and classification of brain tumor using rough K means algorithm and multi kernel SVM in MR images. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-02300-8
Luque-Baena RM, Urda D, Subirats JL, Franco L, Jerez JM (2013) Analysis of cancer microarray data using constructive neural networks and genetic algorithms. In: 2013 Proceedings of the IWBBIO, int work-conference on bioinform and biomed eng, pp 55–63
Mandal S, Banerjee I (2015) Cancer classification using neural network. Int J Emerg Eng Res Technol 3(7):172–178
Mitra P, Mitra S, Pal SK (2000) Staging of cervical cancer with soft computing. IEEE Trans Biomed Eng 47(7):934–940
Mohan A, Rao MD, Sunderrajan S, Pennathur G (2014) Automatic classification of protein structures using physicochemical parameters. Interdiscip Sci Comput Life Sci 6(3):176–186
Qu Y, Ostrouchov G, Samatova N, Geist A (2002) Principal component analysis for dimension reduction in massive distributed data sets. In: 2002 Proceedings of IEEE int conf on data mining (ICDM), pp 1784–1788
Sahu B, Mishra D (2012) A novel feature selection algorithm using particle swarm optimization for cancer microarray data. Procedia Eng 1(38):27–31
Sasikala S, Alias Balamurugan SA, Geetha S (2015) A novel feature selection technique for improved survivability diagnosis of breast cancer. Procedia Comput Sci 50:16–23
Seah CS, Kasim S, Md. Fudzee MF, Hassan R (2020) Significant directed walk framework to increase the accuracy of cancer classification using gene expression data. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-02404-1
Shen Q, Shi WM, Kong W (2008) Hybrid particle swarm optimization and tabu search approach for selecting genes for tumor classification using gene expression data. Comput Biol Chem 32(1):53–60
Stone JV (2002) Independent component analysis: an introduction. Trends Cogn Sci 6(2):59–64
Tarle B, Chintakindi S, Jena S (2020) Integrating multiple methods to enhance medical data classification. Evol Syst 11(1):133–142
Xiao L (2010) A clustering algorithm based on artificial fish school. In: 2010 2nd int conf on comput eng and technol, pp 776–769
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Prakash, P.N.S., Rajkumar, N. Improved local fisher discriminant analysis based dimensionality reduction for cancer disease prediction. J Ambient Intell Human Comput 12, 8083–8098 (2021). https://doi.org/10.1007/s12652-020-02542-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-020-02542-6