Abstract
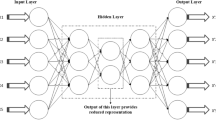
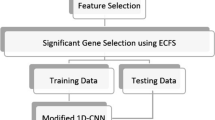
Around the world, cancer is one of the leading reasons of mortality. The importance of earlier detection and prognosis of cancer types is highly significant for patients’ health. In recent research, deep neural networks were trained using gene expression microarray, to classify cancer. Biologists are able to monitor thousands of genes in one experiment using microarray technology. Microarray datasets are considered high-dimensional data, as they are cluttered with irrelevant, redundant, and noisy genes that contribute insignificantly to classification. The most informative genes contributing to cancer classification have been identified using computational intelligence algorithms. In this paper, we propose an integrated framework for cancer classification. This framework is divided into three tasks. Firstly, particle swarm optimization with ensemble learning (PSO-ensemble) reduces the microarray dataset's high dimensionality. Secondly, The Adaptive self-training method (ASTM) is used to solve low-size issues. Finally, a Convolutional Neural Network (CNN) was employed for classification. CNN has the ability to discover the complex non-linear relationships between features and select the most informative. Transfer learning was used sequentially with CNN to integrate the classification procedure because it can reduce the training time and computational complexity. Six microarray datasets are used, namely liver, breast, colon, prostate, central nervous system, and lung. The proposed CNN architecture with transfer learning provided 100% classification accuracy for colon, prostate, CNS and lung microarray datasets, and 97.62%, 95.45% accuracy for liver and breast cancer respectively. Experiments show that our proposed method delivers the highest classification accuracy and reduces training time with the smallest gene subset.





Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Availability of data and materials
Data are available upon request.
Code availability
Code is available upon request.
References
Adem K (2020) Diagnosis of breast cancer with stacked Autoencoder and subspace KNN. Physica A 551:124591. https://doi.org/10.1016/j.physa.2020.124591
Adem K, Kiliçarslan S, Cömert O (2019) Classification and diagnosis of cervical cancer with stacked Autoencoder and Softmax classification. Expert Syst Appl 115:557–564. https://doi.org/10.1016/j.eswa.2018.08.050
Almugren N, Alshamlan H (2019) A Survey on hybrid feature selection methods in microarray gene expression data for cancer classification. IEEE Access 7:78533–78548. https://doi.org/10.1109/ACCESS.2019.2922987
Al-Rajab M, Joan Lu, Qiang Xu (2017) Examining applying high performance genetic data feature selection and classification algorithms for colon cancer diagnosis. Comput Methods Programs Biomed 146:11–24. https://doi.org/10.1016/j.cmpb.2017.05.001
Alrefai N (2019) Ensemble machine learning for leukemia cancer diagnosis based on microarray datasets. Int J Appl Eng Res 14(21):4077–4084
Alrefai N, Ibrahim O (2021a) Semi-supervised ensemble learning for expanding the low sample size of microarray dataset. In: 2021 International conference on electrical, computer and energy technologies (ICECET), IEEE, pp 1–6
Alrefai N, Ibrahim O (2021b) Deep learning-based cancer classification for microarray data: a systematic review. J Theor Appl Inf Technol 99(10):2312–2332. https://doi.org/10.5281/zenodo.6126510
Alrefai N, Ibrahim O (2022) Optimized feature selection method using particle swarm intelligence with ensemble learning for cancer classification based on microarray datasets. Neural Comput Appl. https://doi.org/10.1007/S00521-022-07147-Y
Ang JC, Mirzal A, Haron H, Hamed HNA (2016) Supervised, unsupervised, and semi-supervised feature selection: a review on gene selection. IEEE/ACM Trans Comput Biol Bioinf 13(5):971–989. https://doi.org/10.1109/TCBB.2015.2478454
Baliarsingh K, Santos SV, Muhammad K, Bakshi S (2019) Analysis of high-dimensional genomic data employing a novel bio-inspired algorithm. Appl Soft Comput J 77:520–532. https://doi.org/10.1016/j.asoc.2019.01.007
Barbachan e Silva M, Narloch PH, Dorn M, Broin PO (2021) Optimisation of cancer status prediction pipelines using bio-inspired computing, pp 442–449. https://doi.org/10.1109/cec45853.2021.9504911.
Basavegowda HS, Dagnew G (2020) Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol 5:22–33. https://doi.org/10.1049/trit.2019.0028
Bouazza SH, Auhmani K, Zeroual A, Hamdi N (2018) Selecting significant marker genes from microarray data by filter approach for cancer diagnosis. Proc Comput Sci 127:300–309. https://doi.org/10.1016/J.PROCS.2018.01.126
Cheng R, Jin Y (2015) A social learning particle swarm optimization algorithm for scalable optimization. Inf Sci 291(C):43–60. https://doi.org/10.1016/j.ins.2014.08.039
Cilia ND, De Stefano C, Fontanella F, Raimondo S, Scotto di Freca A (2019) An experimental comparison of feature-selection and classification methods for microarray datasets. Information (switzerland) 10(3):1–13. https://doi.org/10.3390/info10030109
Dabba A, Tari A, Meftali S (2020) Hybridization of moth flame optimization algorithm and quantum computing for gene selection in microarray data. J Ambient Intell Humaniz Comput. https://doi.org/10.1007/s12652-020-02434-9
Dong X, Zhou Y, Wang L, Peng J, Lou Y, Fan Y (2020) Liver Cancer detection using hybridized fully convolutional neural network based on deep learning framework. IEEE Access 8:129889–129898. https://doi.org/10.1109/ACCESS.2020.3006362
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 144(8):1941–1953. https://doi.org/10.1002/ijc.31937
George B, Gokhale SD, Yaswanth PM, Vijayan A, Devika S, Suchithra TV (2022) Identification of Alzheimer associated differentially expressed gene through microarray data and transfer learning-based image analysis. Neurosci Lett 766:136357. https://doi.org/10.1016/J.NEULET.2021.136357
Hengpraprohm S, Jungjit S (2020) Ensemble feature selection for breast cancer classification using microarray data. Intel Artif 23(65):100–114. https://doi.org/10.4114/intartif.vol23iss65pp100-114
Herath HMKKMB, Mittal M (2022) Adoption of artificial intelligence in smart cities: a comprehensive review. Int J Inf Manag Data Insights 2(1):100076. https://doi.org/10.1016/j.jjimei.2022.100076
Hussain S, Muhammad S, Iqbal J, Ahmad I (2020) Optimized gene selection and classification of cancer from microarray gene expression data using deep learning. Neural Comput Appl. https://doi.org/10.1007/s00521-020-05367-8
Kim S, Park J (2018) Hybrid feature selection method based on neural networks and cross-validation for liver cancer with microarray. IEEE Access 6:78214–78224. https://doi.org/10.1109/ACCESS.2018.2884896
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97(1–2):273–324. https://doi.org/10.1016/S0004-3702(97)00043-X
Kumar CA, Ramakrishnan S (2015) Binary classification of cancer microarray gene expression data using extreme learning machines. In: 2014 IEEE International Conference on computational intelligence and computing research, IEEE ICCIC 2014, pp 29–38, https://doi.org/10.1109/ICCIC.2014.7238297
Liao Q, Jiang L, Wang X, Zhang C, Ding Y (2017) Cancer classification with multi-task deep learning. In: 2017 International conference on security, pattern analysis, and cybernetics (SPAC). IEEE, pp 76–81
Liu J, Bolei Xu, Zheng C, Gong Y, Garibaldi J, Soria D, Green A, Ellis IO, Zou W, Qiu G (2019) An end-to-end deep learning histochemical scoring system for breast cancer TMA. IEEE Trans Med Imaging 38(2):617–628. https://doi.org/10.1109/TMI.2018.2868333
Mazumder DH, Veilumuthu R (2019) An enhanced feature selection filter for classification of microarray cancer data. ETRI J 41(3):358–370. https://doi.org/10.4218/etrij.2018-0522
Medjahed SA, Saadi TA, Benyettou A, Ouali M (2017) Kernel-based learning and feature selection analysis for cancer diagnosis. Appl Soft Comput J 51:39–48. https://doi.org/10.1016/j.asoc.2016.12.010
Molina D, Poyatos J, Del Ser J, García S, Hussain A, Herrera F (2020) Comprehensive taxonomies of nature- and bio-inspired optimization: inspiration versus algorithmic behavior, critical analysis recommendations. Cogn Comput. https://doi.org/10.1007/s12559-020-09730-8
Nilashi M, bin Ibrahim O, Ahmadi H, Shahmoradi L (2017) An analytical method for diseases prediction using machine learning techniques. Comput Chem Eng 106:212–223. https://doi.org/10.1016/J.COMPCHEMENG.2017.06.011
Oza NC, Tumer K (2008) Classifier ensembles: select real-world applications. Inf Fusion 9(1):4–20. https://doi.org/10.1016/J.INFFUS.2007.07.002
Panda M (2018) Elephant search optimization combined with deep neural network for microarray data analysis. J King Saud Univ Comput Inf Sci 32(8):940–948. https://doi.org/10.1016/j.jksuci.2017.12.002
Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JYH, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR (2002) Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415(6870):436–442. https://doi.org/10.1038/415436a
Rahman J, Ahammed B, Abedin M, Suri JS (2019) Computer methods and programs in biomedicine statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput Methods Programs Biomed 176:173–193. https://doi.org/10.1016/j.cmpb.2019.04.008
Salem H, Attiya G, El-fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput J 50:124–134. https://doi.org/10.1016/j.asoc.2016.11.026
Sarkar C, Cooley S, Srivastava J (2014) Robust feature selection technique using rank aggregation. Appl Artif Intell 28(3):243–257. https://doi.org/10.1080/08839514.2014.883903
Shen Q, Diao R, Su P (2012) Feature selection ensemble. Turing-100 10:289–306. https://doi.org/10.29007/rlxq
WHO (2020) Cancer. World Health Organization. https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 23 June 2021
Xia C, Yawen X, Jun W, Xiaodong Z, Hua L (2019) A convolutional neural network based ensemble method for cancer prediction using DNA methylation data, pp 191–96. https://doi.org/10.1145/3318299.3318372.
Xu B, Liu J, Garibaldi J, Ellis IO, Soria D, Gong Y, Zheng C, Green A, Qiu G, Zou W (2019) An end-to-end deep learning histochemical scoring system for breast cancer TMA. IEEE Trans Med Imaging 38(2):1–1. https://doi.org/10.1109/tmi.2018.2868333
Zhu Z, Ong Y-S, Dash M (2007) Markov Blanket-embedded genetic algorithm for gene selection. Pattern Recogn 40(11):3236–3248. https://doi.org/10.1016/j.patcog.2007.02.007
Funding
This study did not receive external or internal funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest that are relevant to the content of this article.
Ethics approval
All information and the data source used in our study were mentioned in the research, and it is available and public for research purposes.
Consent for publication
We used a public dataset and cited it appropriately in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alrefai, N., Ibrahim, O., Shehzad, H.M.F. et al. An integrated framework based deep learning for cancer classification using microarray datasets. J Ambient Intell Human Comput 14, 2249–2260 (2023). https://doi.org/10.1007/s12652-022-04482-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12652-022-04482-9